More publications
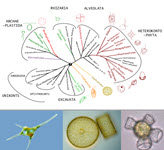
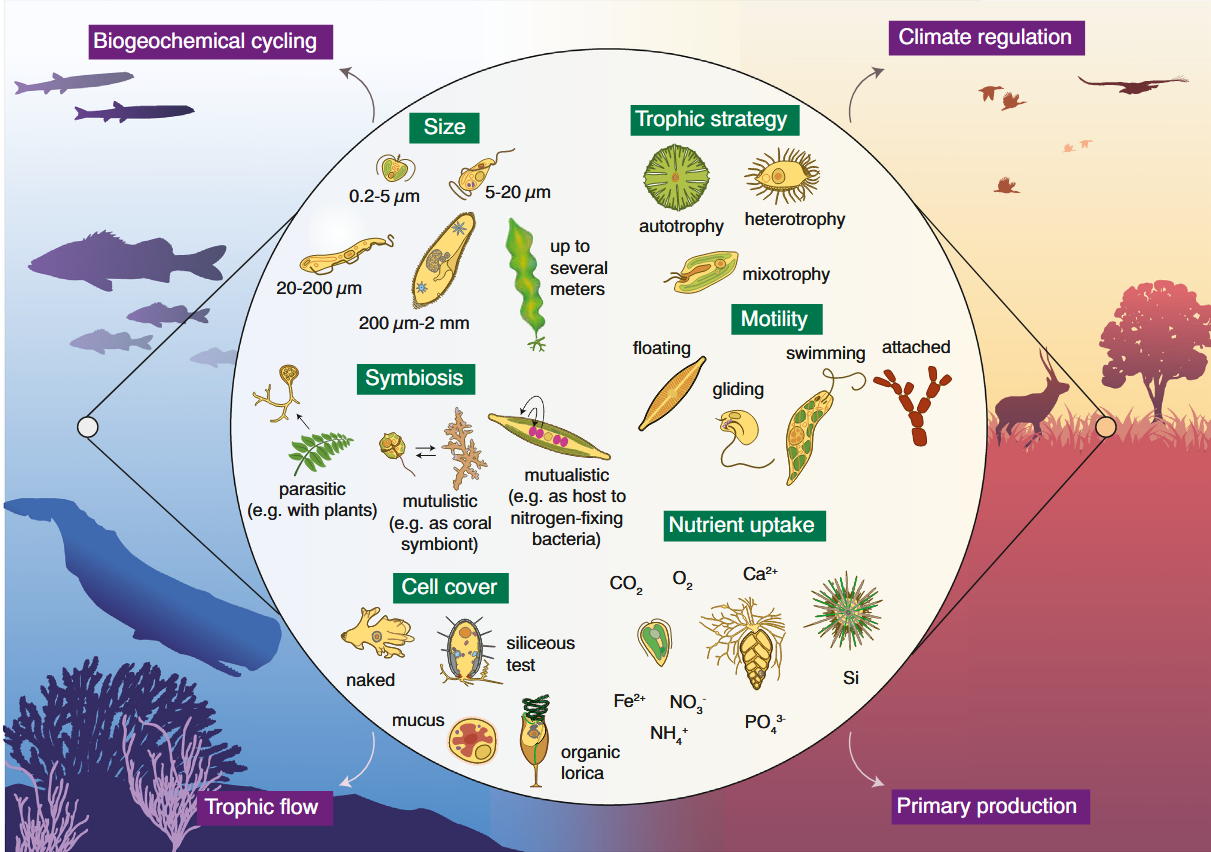
We argue that a unified trait database for protists is timely, achievable, and would catalyze transformative research on their biodiversity, ecology and evolution.
Mahwash Jamy, Pierre Ramond, David Bass, Javier Del Campo, Micah Dunthorn, Enrique Lara, Aditee Mitra, Daniel Vaulot, Luciana Santoferrara
Trends in Microbiology,
2025
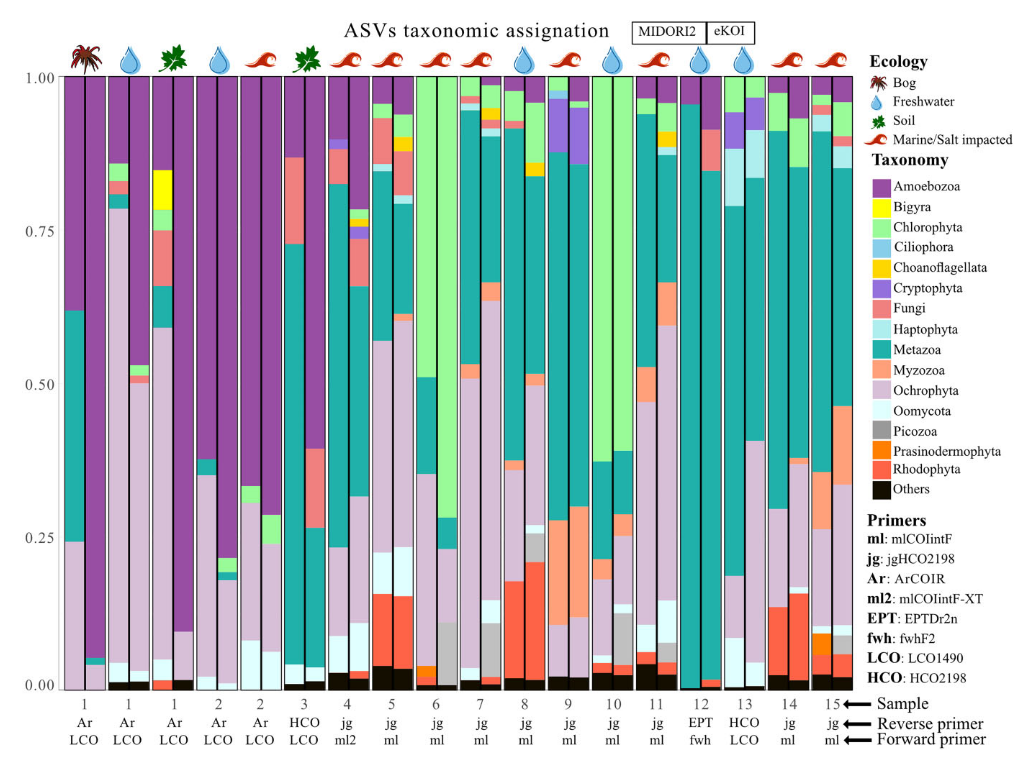
We introduce eKOI, a novel, curated COI gene database designed to enhance the taxonomic annotation for protists that can be used for COI-based metabarcoding.
Rubén González-Miguéns, Àlex Gàlvez-Morante, Margarita Skamnelou, Meritxell Antó, Elena Casacuberta, Daniel J Richter, Enrique Lara, Daniel Vaulot, Javier del Campo, Iñaki Ruiz-Trillo
ISME Communications, 5: ycaf075,
2025
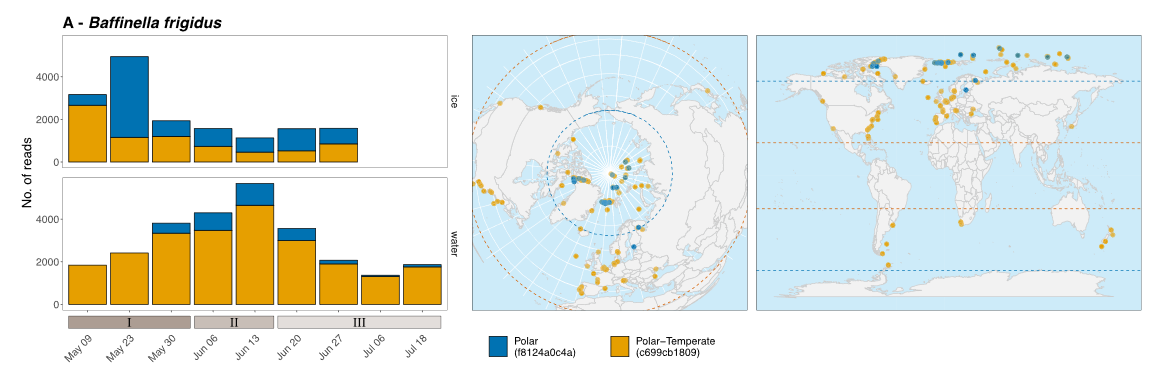
We analyzed the phytoplankton dynamics during the under ice bloom both in the water and in the ice off Baffin island (Canada).
Clarence Wei Hung Sim, Catherine Ribeiro Gerikas, Florence Le Gall, Ian Probert, Priscilla Gourvil, Connie Lovejoy, Daniel Vaulot, Adriana Lopes dos Santos
ISME Communications, 5: ycaf075,
2025
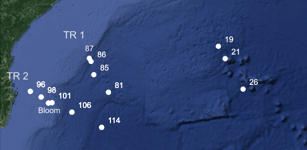
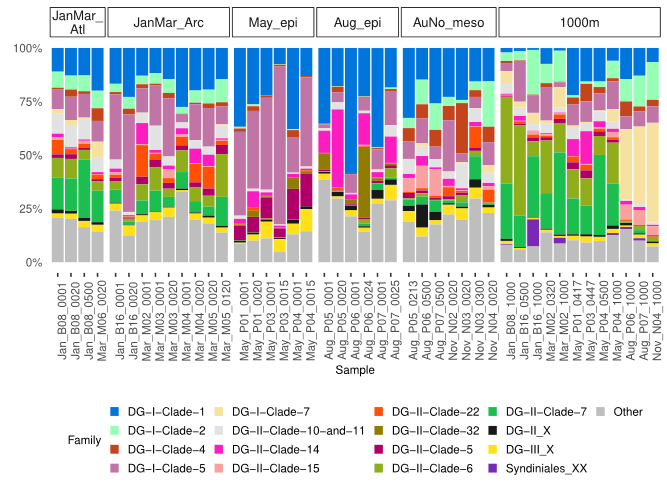
We investigated the seasonal variability marine alveolate community structure in the Arctic Ocean west and north of Svalbard.
Elianne Egge, Daniel Vaulot, Aud Larsen, Bente Edvardsen
Arctic Science 11:1-16,
2025
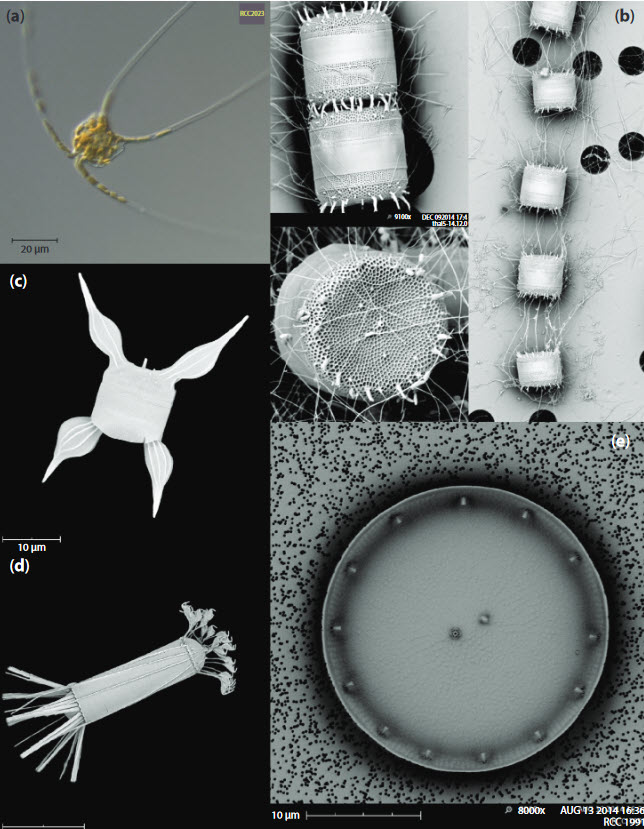
A review on diatom culture.
Daniel Vaulot, Gust Bilcke, Peter Chaerle, Angela Falcatiore, Priscillia Gourvil, Michael W. Lomas, Ian Probert, Wim Vyverman
In Goessling, J., Serodio, J. & Lavaud, J. [Eds.] Diatom Photosynthesis: From Primary Production to High-Value Molecules. Wiley-Scrivener, pp. 409–48.,
2024
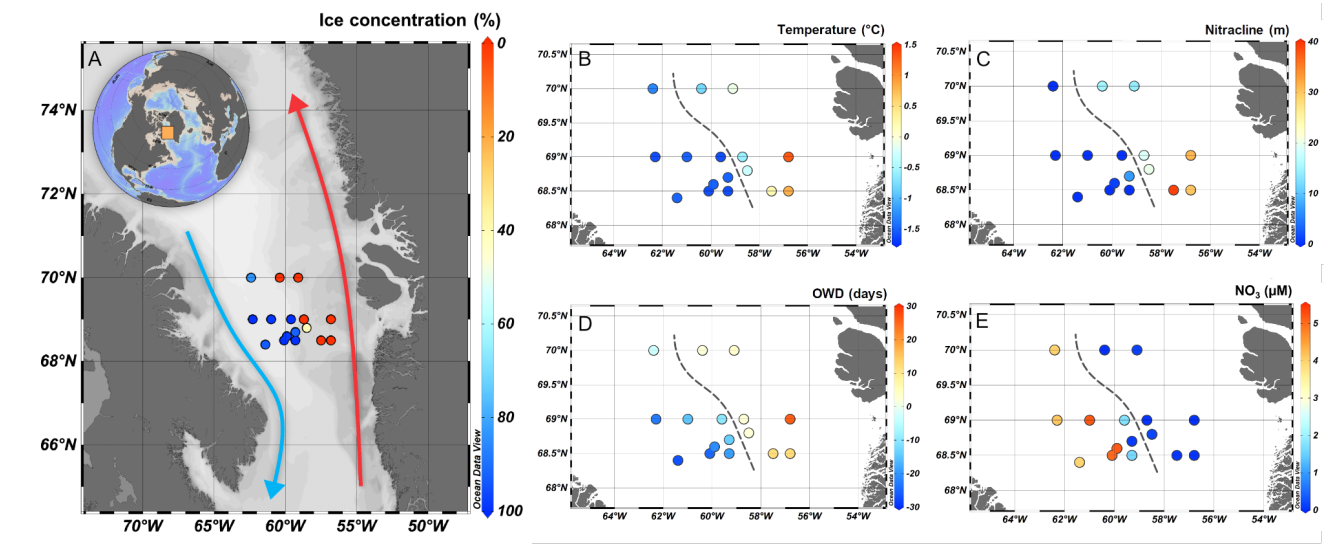
We investigated the phytoplankton community structure across the marginal ice zone between the ice-free, Atlantic-influenced, east and the ice-covered, Arctic-influenced, west Baffin Bay using 18S rRNA high-throughput amplicon sequencing.
Catherine Ribeiro Gerikas, Adriana Lopes dos Santos, Nicole Trefault, Dominique Marie, Connie Lovejoy, Daniel Vaulot
Elementa: Science of the Anthropocene,
2024