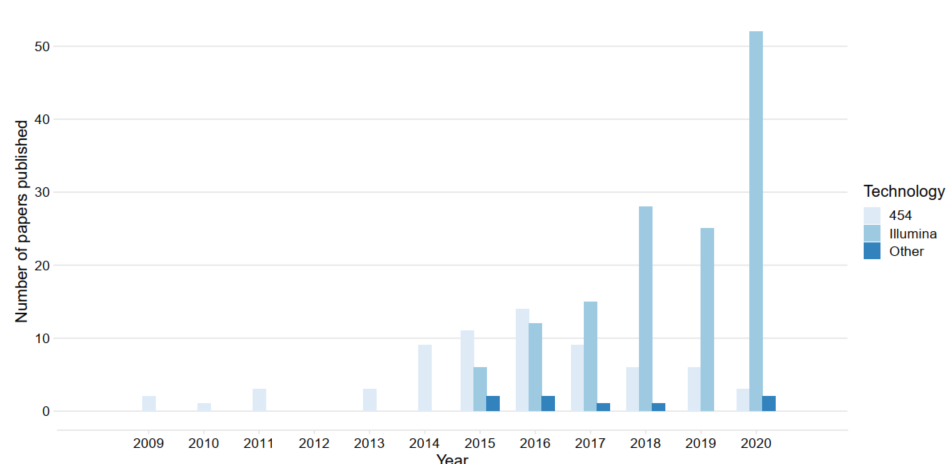
 Samples from studies using metabarcoding for aquatic protist studies
Samples from studies using metabarcoding for aquatic protist studies
Phytoplankton diversity and ecology through the lens of high throughput sequencing technologies
Abstract
Metabarcoding or high-throughput sequencing of a specific genetic marker is a powerful technique, widely used today, to analyze biodiversity across distinct environments and taxonomic groups. Plankton ecologists have benefited tremendously from the growing accumulation of metabarcoding studies. Novel biogeographic patterns have been established for cyanobacteria and microbial eukaryotes by the analysis of datasets from the Tara Oceans and Ocean Sampling Day projects. Novel lineages without cultured representatives have been uncovered. This chapter begins by going back to Carl Woese and George Fox who were the first to define the concept of “molecular marker”. We then detail the different steps and choices that are involved in designing, performing and analysing a metabarcoding study. We are using a compilation of about 250 metabarcoding studies to present the major trends in terms of gene markers used and environments probed. We are then focusing on specific habitats and processes that have benefited from metabarcoding: the study of polar ecosystems, the functioning of the marine biological carbon pump, predator-prey interactions, and picoeukaryotic phytoplankton in highly urbanized lakes. An alternative approach to metabarcoding developed for marine picocyanobacteria is also briefly discussed. Finally, we offer some perspectives on emerging trends such as the use of metabarcodes combined with supervised machine learning for bio-monitoring, the link between metabarcoding and functional diversity in trait-based studies and the massive sequencing of long DNA fragments