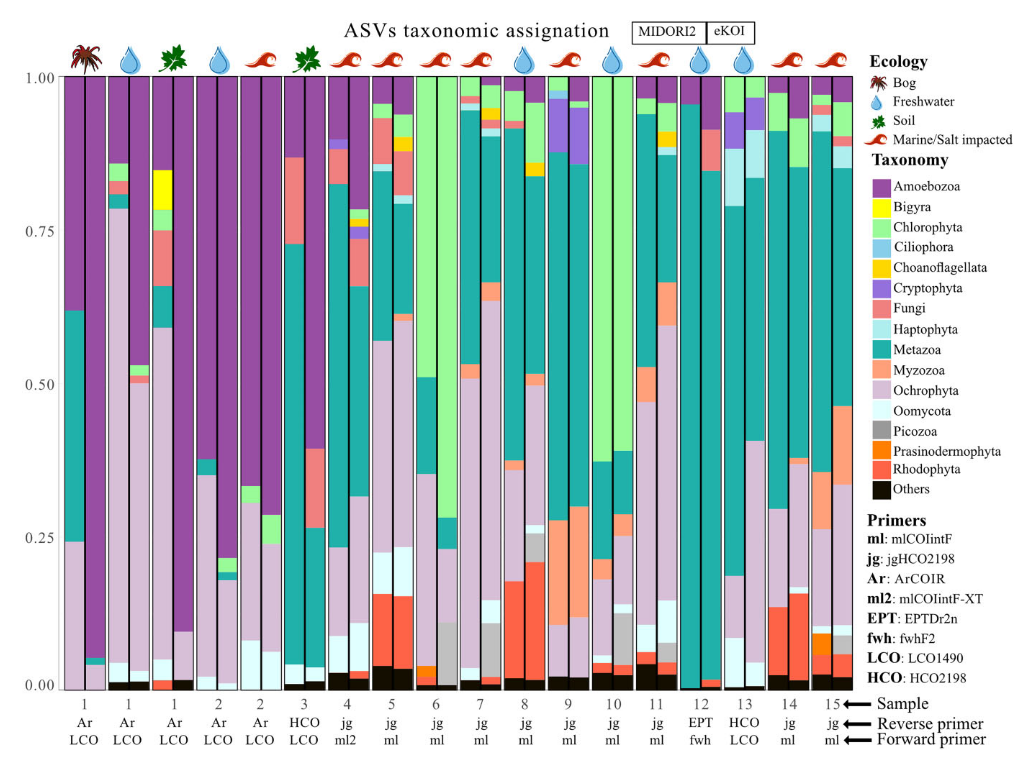
 Improved annotation with eKOI database.
Improved annotation with eKOI database.
A novel taxonomic database for eukaryotic mitochondrial cytochrome oxidase subunit I gene (eKOI), with a focus on protists diversity
Abstract
Metabarcoding has emerged as a robust method for assessing biodiversity patterns by retrieving environmental DNA directly from ecosystems. While the 18S rRNA gene is the primary genetic marker used for broad eukaryotic metabarcoding, it has limitations in resolving lower taxonomic levels. A potential alternative is the mitochondrial cytochrome oxidase subunit I (COI) gene because it offers resolution at the species level. However, the COI gene lacks a comprehensive, curated taxonomically informed database including protists. To address this gap, we introduce eKOI, a novel, curated COI gene database designed to enhance the taxonomic annotation for protists that can be used for COI-based metabarcoding. eKOI integrates data from GenBank and mitochondrial genomes, followed by extensive manual curation to eliminate redundancies and contaminants, recovering 15 947 sequences within 80 eukaryotic phyla. We validated the use of eKOI by reannotating several COI metabarcoding datasets, revealing previously unidentified protist biodiversity and demonstrating the database utility for community-level analyses.