Daniel Vaulot
2024-01-24

Introduction to Roscoff ABIMS server
Outline
- Pre-requisites
- Connecting to server
- Linux basics
- Running programs - SLURM
Pre-requisites
Software
Terminal program
File transfer
File editing
Reference
Linux
ABIMS Roscoff cluster
SLURM (job management)
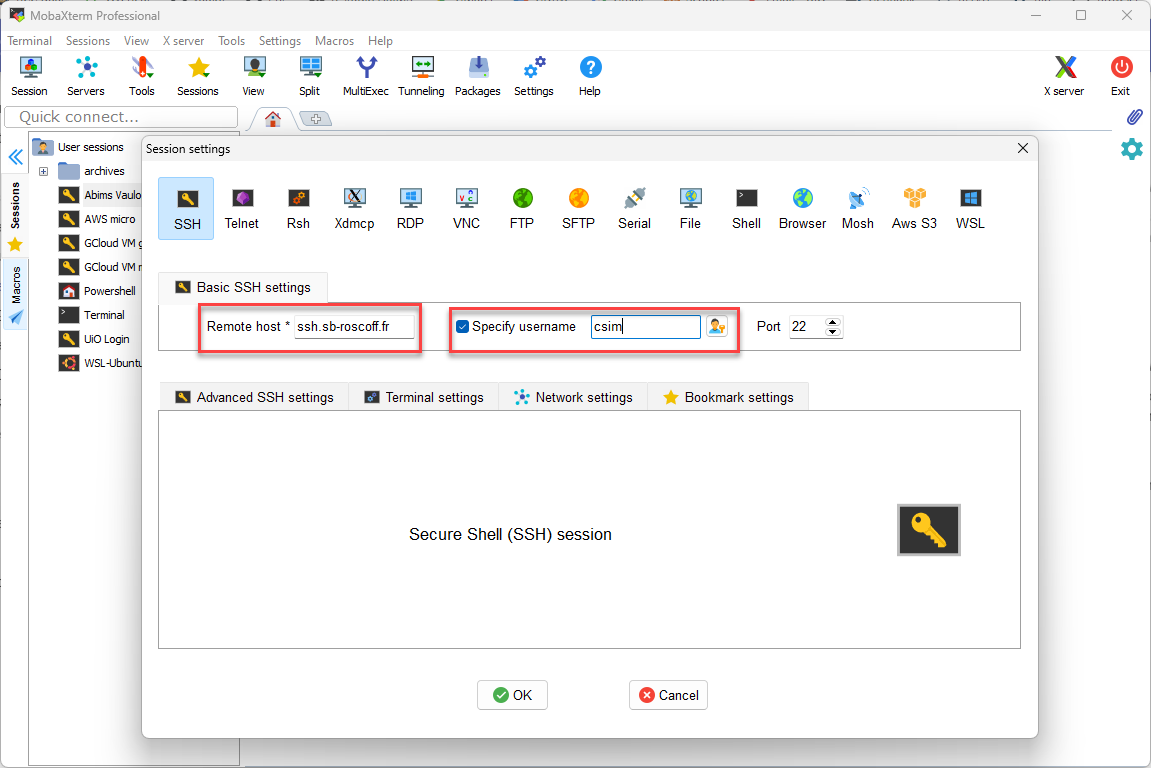
Connecting to server
Connecting to ABIMS terminal
Launch MobaXterm
Enter information in new session
Connecting to ABIMS terminal
- Type password (cannot copy and paste)
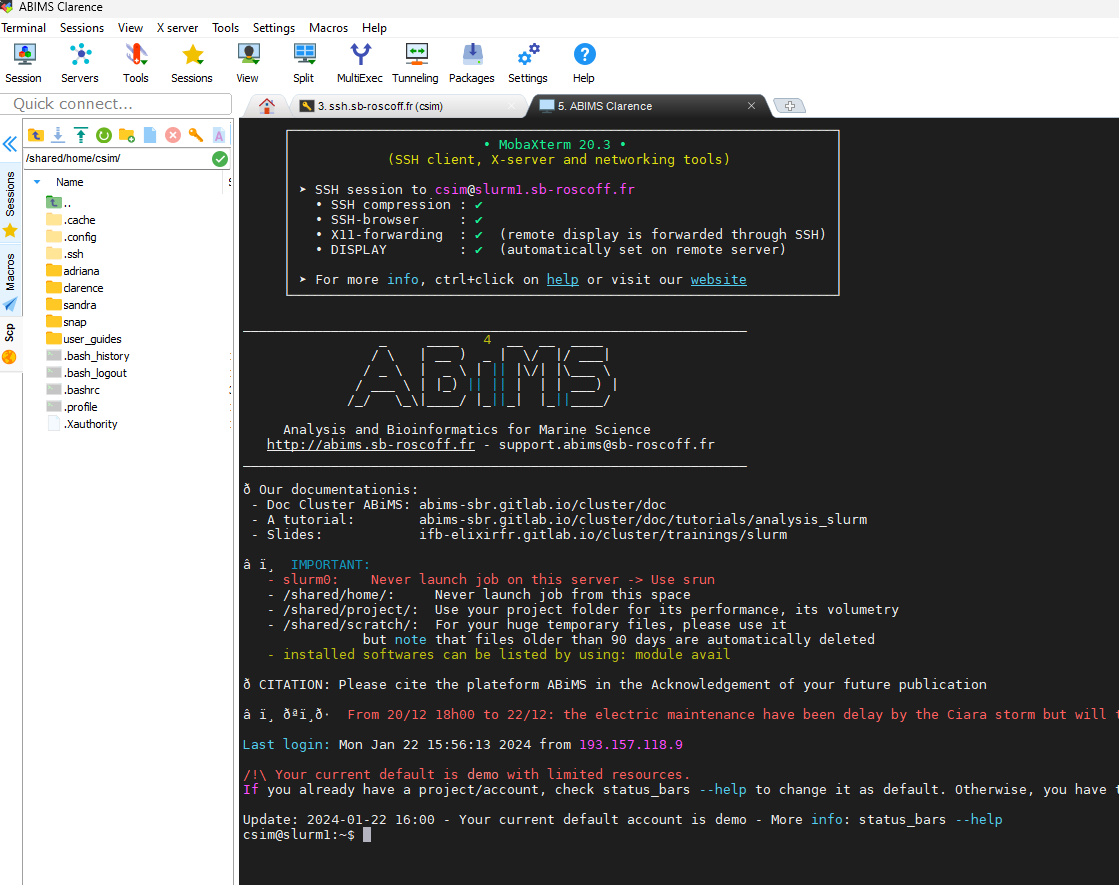
Connecting to ABIMS terminal
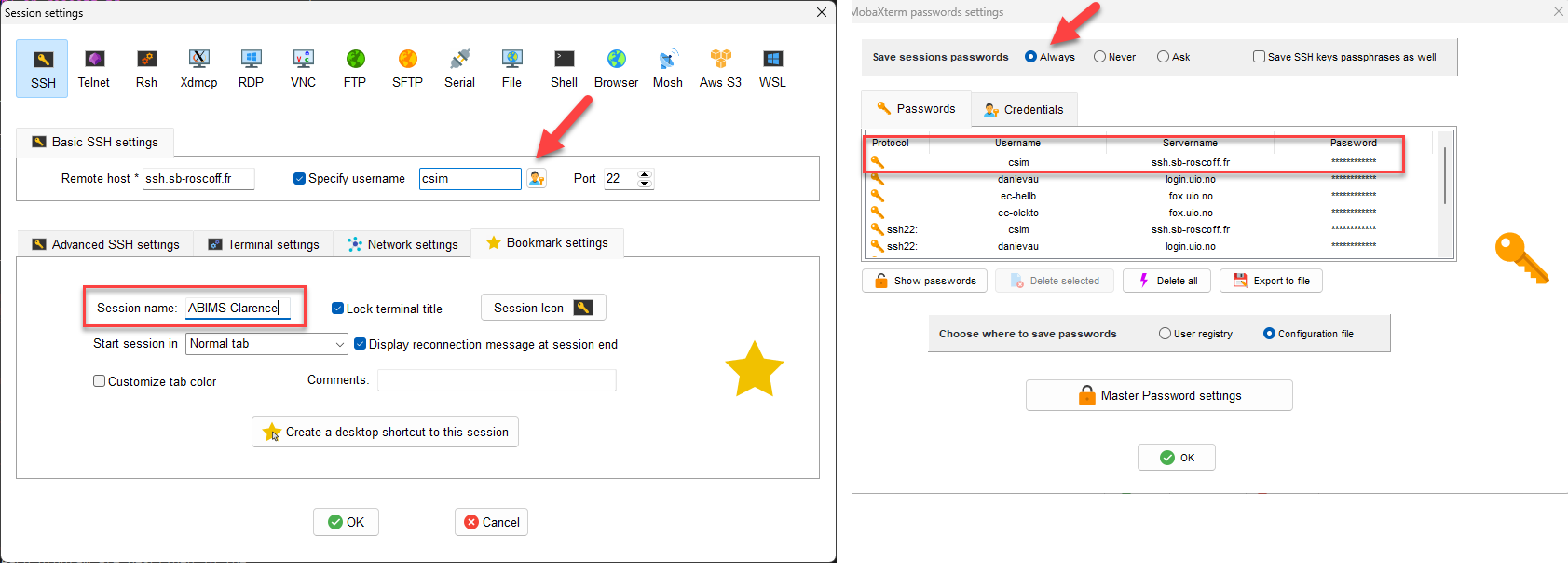
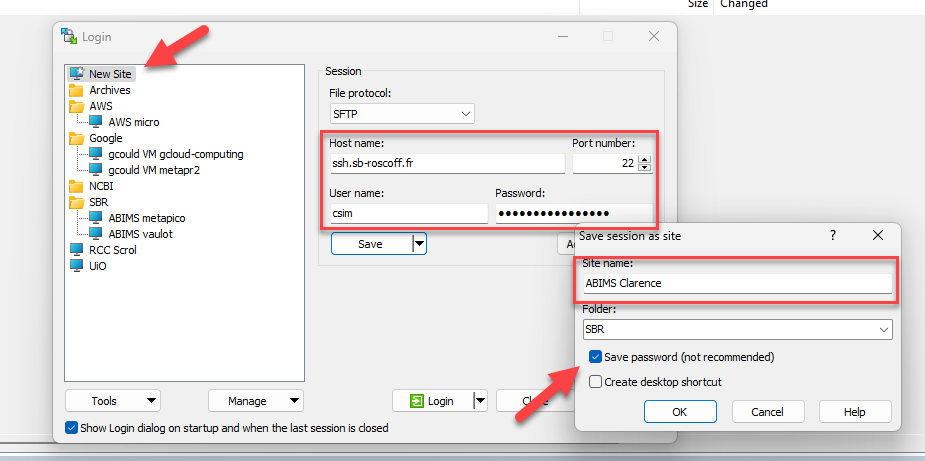
Rename session
The password is saved in the session (save passwords = always)
Upload/Download files system with WinSCP
Launch WinSCP
Create new site
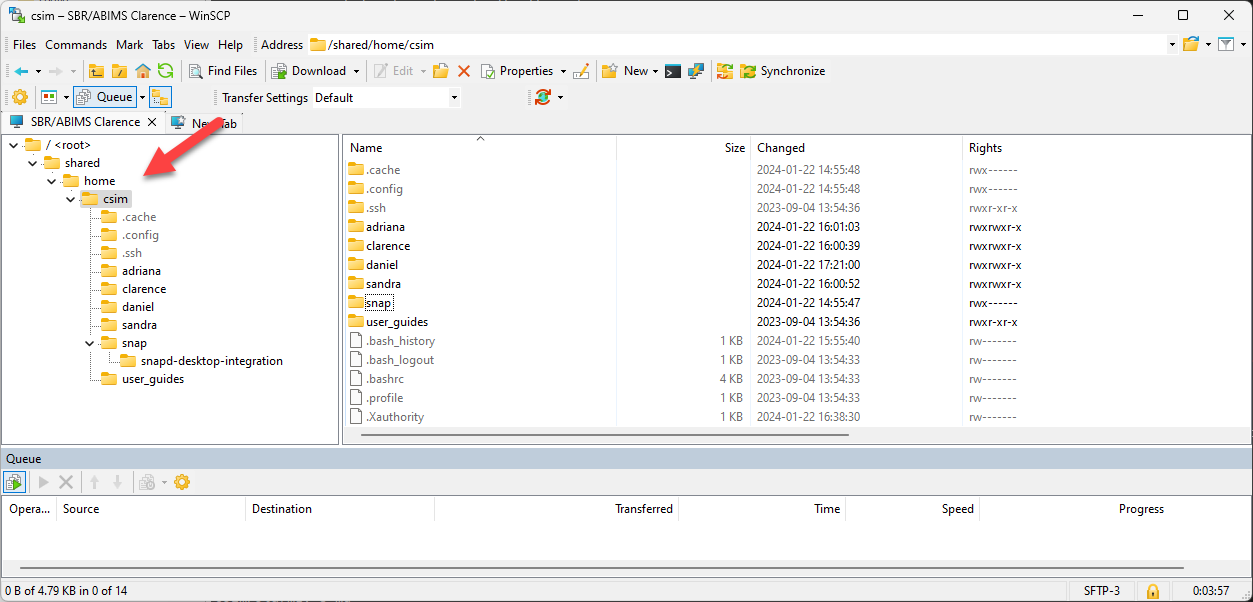
Navigate the file system
Exit system
Intro to Linux
Very important points
- Linux is case sensitive
- No space into file names, always cause problem
- Directories are separated by “/” (not “" like in Windows)
- Keep everything organized because Linux programs create lots of output files
Navigate the file system
- Where am I ? pwd: present working directory
- You are in the home directory of csim
Navigate the file system
- Change directory: cd
Create and manipulate files
Create files using winscp
- Update preferences to make VS Code default editor
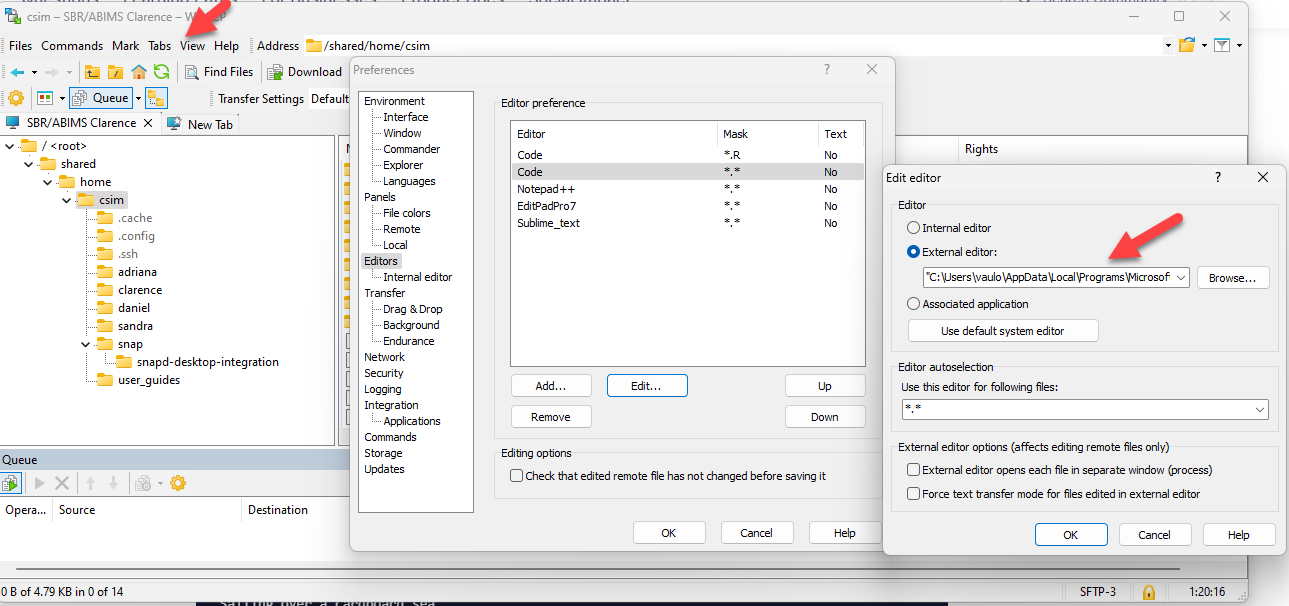
- Create new file in your directory
- Double click to edit
- Save under VS Code and it will be updated in server
Display file directly in terminal
Tips and tricks
- Up and Down arrow call back previous lines
- Use TAB to finish a command
- Copy - Paste (do not use CTR-C, CTR-V)
- Copy: just select the part you want to copy
- Paste: Right click on mouse
Tips and tricks
Pipes
- “>” Redirect output towards a file
Tips and tricks
Variables
Job management (SLURM)
Motivation
- Programs
- Run for long (cannot wait for output)
- Need specific amount of memory
- Can run on multiple processors
- Many user simultaneously
- Examples of programs
- R
- mafft: alignments
- raxml: trees
- vsearch: clustering, sequence manipulation
- emu: metabarcode assignment
- Must always be located in project folder
Move to project folder - ALWAYS
Our project are located in /shared/projects/geek_simple_laby
- Only three folders are backed up.
- You can copy your scripts to the
scriptfolder under your directory (script/sandra). - Always do your own backup on your PC (download files/folder)
# Project working directory usage at ABiMS
ABiMS provides a backup on your project directory.
To take advantage of this process, you have to follow some rules:
- Only the subdirectories ‘archive’, ‘script’ and ‘finalresult’ are backed up.
- You must place these subdirectories at the root of your project folder.
- Please be smart in your backups for our finances and the planet.Interactive mode - srun
- Use for programs like R
- Not really recommended
- Do not forget to quit when done (CPU time)
Batch mode - sbatch
- Copy the necessary files
What we are going to do:
- Singapore time series ASVs for Labyrinthulomycetes
- 289 sequences, but some are very similar
- Clusterize using 99% similarity
- Use vsearch program
- Final: 78 sequences
Input file
>pacbio;3d4a51f4d1;Aplanochytrium_sp.;size=120;
AGCTCCAATAGCGTATATTAAAGTTGTTGCAGTTAAAAAGCTCGTAGTTGGATTTCTGGTAGGAGCGACCGTGCCGAACTTGATTGTTCGTGTATTGTGTTGTCTTCAGCCATCCTCGT
GGAGAACTTTTCTAACATTAACTTGTTGGGATTGGGACCCGCGTCGTTTACTGTGAAAAAATTAGAGTGTTTAAAGCAGGCATTAGCTTGAATACATTAGCATGGAATAATAAGATAGG
ACTTTGGTACTATTTTGTTGGTTTGCATACCAAATTAATGATCAACAGGAACAGTTTGAGGATATTCGTATGAACATGTCAGAGGTGAAATTCTTGGATTTTGATCAGACGAACTACTG
CGAAAGCATTTATCAAGGATGTTTTCATTAATCAAGAACGAAAGTTAGGGGATCGAA...Batch mode - sbatch
sbatch_cluster_01.sh
Two parts:
- Header (very strict formatting)
- Command to Run
#!/bin/bash
#SBATCH -p fast # Partition can be also fast, long, bigmem
#SBATCH --cpus-per-task 4
#SBATCH --mem-per-cpu 4GB # mémoire vive pour l'ensemble des cœurs
#SBATCH -t 6-0:00 # durée maximum du travail (D-HH:MM)
#SBATCH -o slurm.%N.%j.out # STDOUT
#SBATCH -e slurm.%N.%j.err # STDERR
#SBATCH --mail-user=vaulot@sb-roscoff.fr # ! Replace with uio email
#SBATCH --mail-type=BEGIN,END,FAIL
# Submitted with
# cd /shared/home/csim/daniel # ! Change to your directory
# sbatch sbatch_cluster_01.sh
module load vsearch
cd /shared/home/csim/daniel # ! Change to your directory
"${VSEARCH}" --cluster_fast "Labyrinthulomycetes.pacbio.fasta" \
--threads 4 \
--id 0.99 \
--uc clusters_0.99_Labyrinthulomycetes.pacbio.tsv \
--sizeout \
--centroids clusters_0.99_Labyrinthulomycetes.pacbio.centroids.fasta \
--clusterout_sort \
--clusterout_idNeed to edit: email and directories
Batch mode - sbatch
Get status of your run
Three states:
- PENDING
- RUNNING
- COMPLETED
squeue
You should also get two emails:
- Slurm Job_id=36263527 Name=sbatch_cluster_01.sh Began, Queued time 00:00:01
- Slurm Job_id=36263527 Name=sbatch_cluster_01.sh Ended, Run time 00:00:23, COMPLETED, ExitCode 0
Batch mode - sbatch
Files produced
slurm.cpu-node-050.36263527.err
slurm.cpu-node-050.36263527.out
clusters_0.99_Labyrinthulomycetes.pacbio.centroids.fasta
clusters_0.99_Labyrinthulomycetes.pacbio.tsv
slurm.cpu-node-050.36263527.err
Output of the program vsearch
vsearch v2.22.1_linux_x86_64, 251.3GB RAM, 256 cores
https://github.com/torognes/vsearch
Reading file Labyrinthulomycetes.pacbio.fasta 100%
1327126 nt in 289 seqs, min 4174, max 5443, avg 4592
Masking 100%
Sorting by length 100%
Counting k-mers 100%
Clustering 100%
Sorting clusters 100%
Writing clusters 100%
Clusters: 78 Size min 1, max 29, avg 3.7
Singletons: 41, 14.2% of seqs, 52.6% of clustersBatch mode - sbatch
clusters_0.99_Labyrinthulomycetes.pacbio.centroids.fasta
>pacbio;1a9244a2e6;Thraustochytriaceae_X_sp.;clusterid=74;size=29
AGCTTCAATAGCATATACTAACGTTGTCGCAGTTAAAAAGTTCGTAGTTGAATTTCTGGTAGGAGTGACCTGGCCTTTTA
CGTTTGTAATTGTATGCTGTGTGTTATCTCTGGCCATCCTGAATCTGCTTTGTTGTAGATTCTCACATACTGTAAAAAAA
TTAGAGTGTTTAAAGCATTTCGTATGAAAAGAATACATCTTATGGGATATCAAAATAGGATTTTGGTGCTATTTTGTTGG
TTTGCACACCAAAATAATGATTAACAGGGACAGTTGGGGGTATTTGTATTTAATTGTCAGAGGTGAAATTCTTGGATTTA
TGAAAGACAAACTACTGCGAAAGCATTTATCAAGGATGTTTTCATTAATCATGAACGAAAGTTAGGGGATCGAAGATGAT
CAGATACCATCGTAGTCTTAACAGTAAACTATACCAACTTGCGATTATTCCATGGTGTTTTTTGCCAGGAGTAGCAGCACclusters_0.99_Labyrinthulomycetes.pacbio.tsv
S 0 5443 * * * * * pacbio;4b0f43a6e4;Labyrinthulomycetes_LAB8_sp.;size=3; *
H 0 5436 99.4 + 0 0 1120MI12M7I673MI289M3D174M2D1534M3I1629M pacbio;e3f28aa0af;Labyrinthulomycetes_LAB8_sp.;size=4; pacbio;4b0f43a6e4;Labyrinthulomycetes_LAB8_sp.;size=3;
H 0 5436 99.4 + 0 0 1120MI12M7I673MI289M3D174M2D1534M3I1629M pacbio;70ee1479b8;Labyrinthulomycetes_LAB8_sp.;size=3; pacbio;4b0f43a6e4;Labyrinthulomycetes_LAB8_sp.;size=3;
S 1 5230 * * * * * pacbio;86192ef156;Labyrinthulomycetes_LAB8_sp.;size=15; *
H 1 5229 99.8 + 0 0 1267MI530MD301MI3130M pacbio;e54d7a85c5;Labyrinthulomycetes_LAB8_sp.;size=4; pacbio;86192ef156;Labyrinthulomycetes_LAB8_sp.;size=15;
H 1 5227 99.5 + 0 0 1120MI12M7I634M3D24MD305MD3127M pacbio;970a95784e;Labyrinthulomycetes_LAB8_sp.;size=7; pacbio;86192ef156;Labyrinthulomycetes_LAB8_sp.;size=15;
H 1 5227 99.5 + 0 0 1120MI12M7I634M3D24MD305MD3127M pacbio;d23a7e7d59;Labyrinthulomycetes_LAB8_sp.;size=2; pacbio;86192ef156;Labyrinthulomycetes_LAB8_sp.;size=15;
H 1 5226 99.7 + 0 0 1267MI555MI275MI39MI3090M pacbio;abfdcca79c;Labyrinthulomycetes_LAB8_sp.;size=9; pacbio;86192ef156;Labyrinthulomycetes_LAB8_sp.;size=15;Batch mode - sbatch
If something goes wrong:
- Always PENDING: Problem with the SLURM instructions at top of file
- Script error check CAREFULLY .err file
Error in file names
slurm.cpu-node-050.36263532.err
Batch mode - sbatch
sbatch_cluster_02.sh
- Replace strings by variables
- Can re-use script for different files and parameters
- Easier to track errors
module load vsearch
DIR="/shared/home/csim/daniel/" # ! Change to your directory
FILE_HEAD="Labyrinthulomycetes.pacbio"
IDENTITY="0.99"
THREADS=4
cd $DIR
vsearch --cluster_fast "${FILE_HEAD}.fasta" \
--threads "${THREADS}" \
--id "${IDENTITY}" \
--uc clusters_${IDENTITY}_$FILE_HEAD.tsv \
--msaout clusters_${IDENTITY}_${FILE_HEAD}.align.fasta \
--sizeout \
--centroids clusters_${IDENTITY}_${FILE_HEAD}.centroids.fasta \
--clusterout_sort \
--clusterout_idMore advanced processing
Loop through files
Loop through parameters
Work directly on files
- grep: search (regular expression)
- sed: edit text
tar/zip/unzip: compress files/rectories
wget: download files from internet