Microbes
Week 11 - Analyzing protist communities
MetaPR2
A database of metabarcodes
Daniel Vaulot
2023-01-17


Outline
Metabarcoding data
Factors affecting protist communities
Diversity
Visualization/Analysis
MetaPR2 in practice
Final presentation
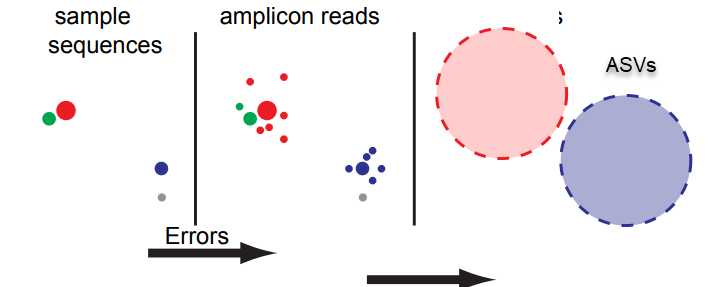
Metabarcoding
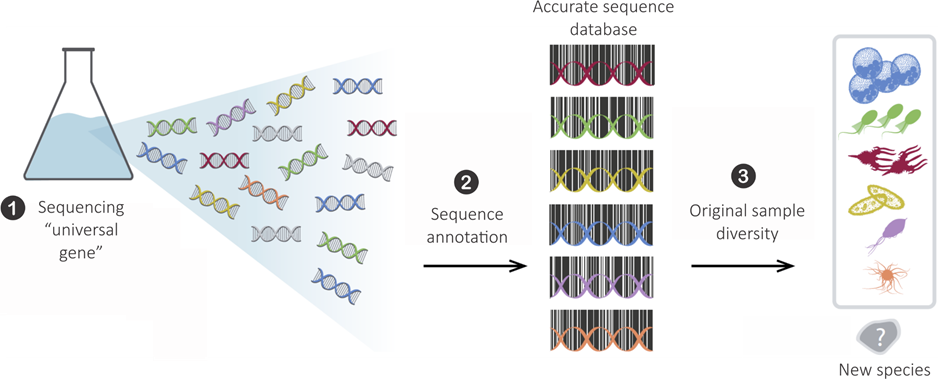
Metabarcoding
Metabarcoding
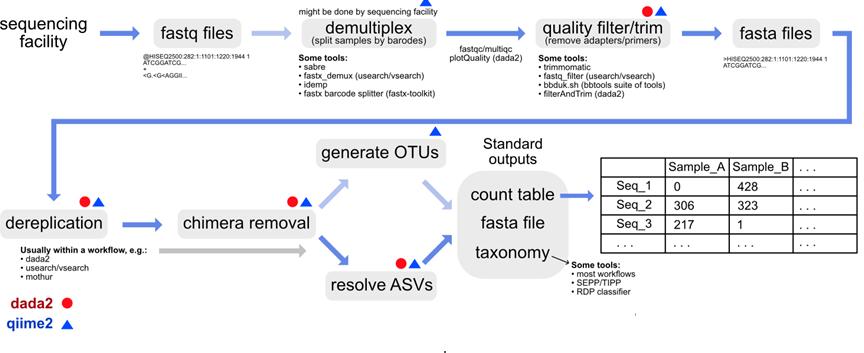
Metabarcoding
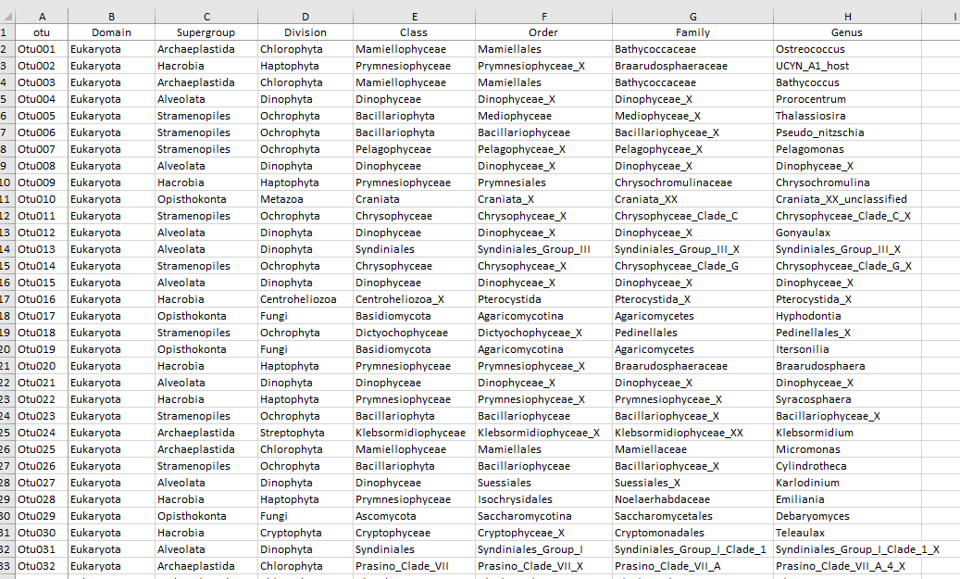
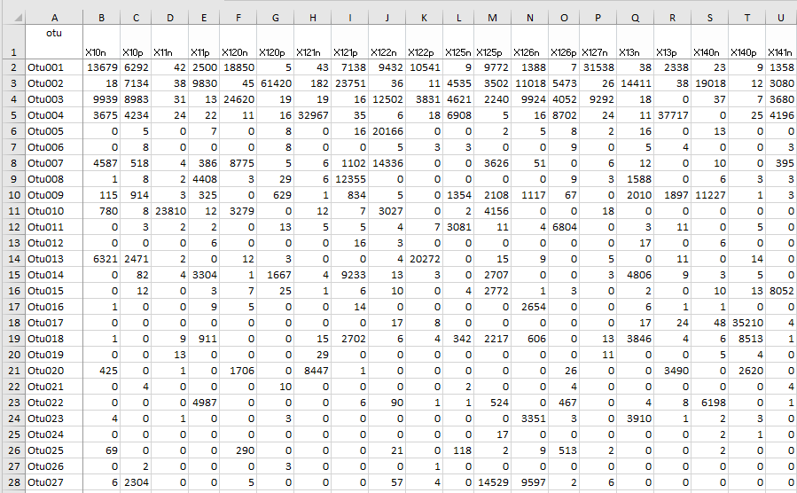
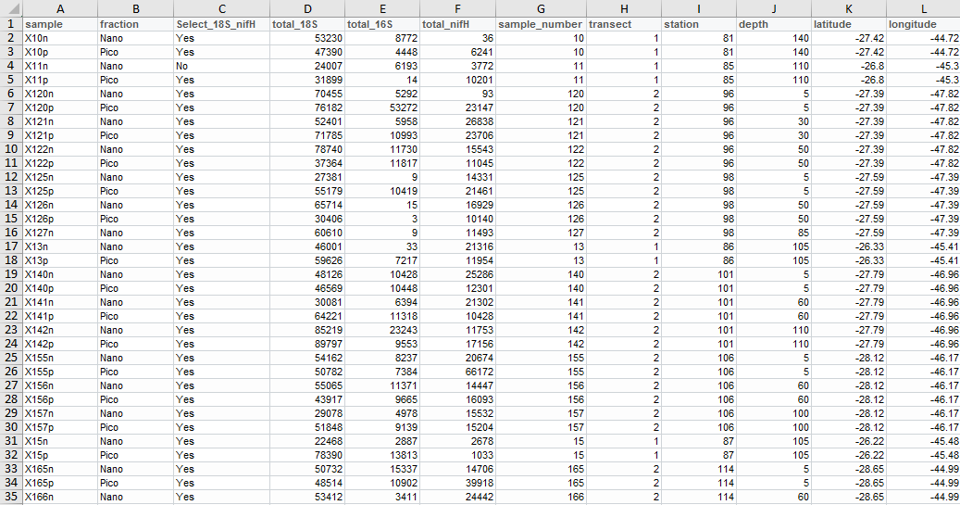
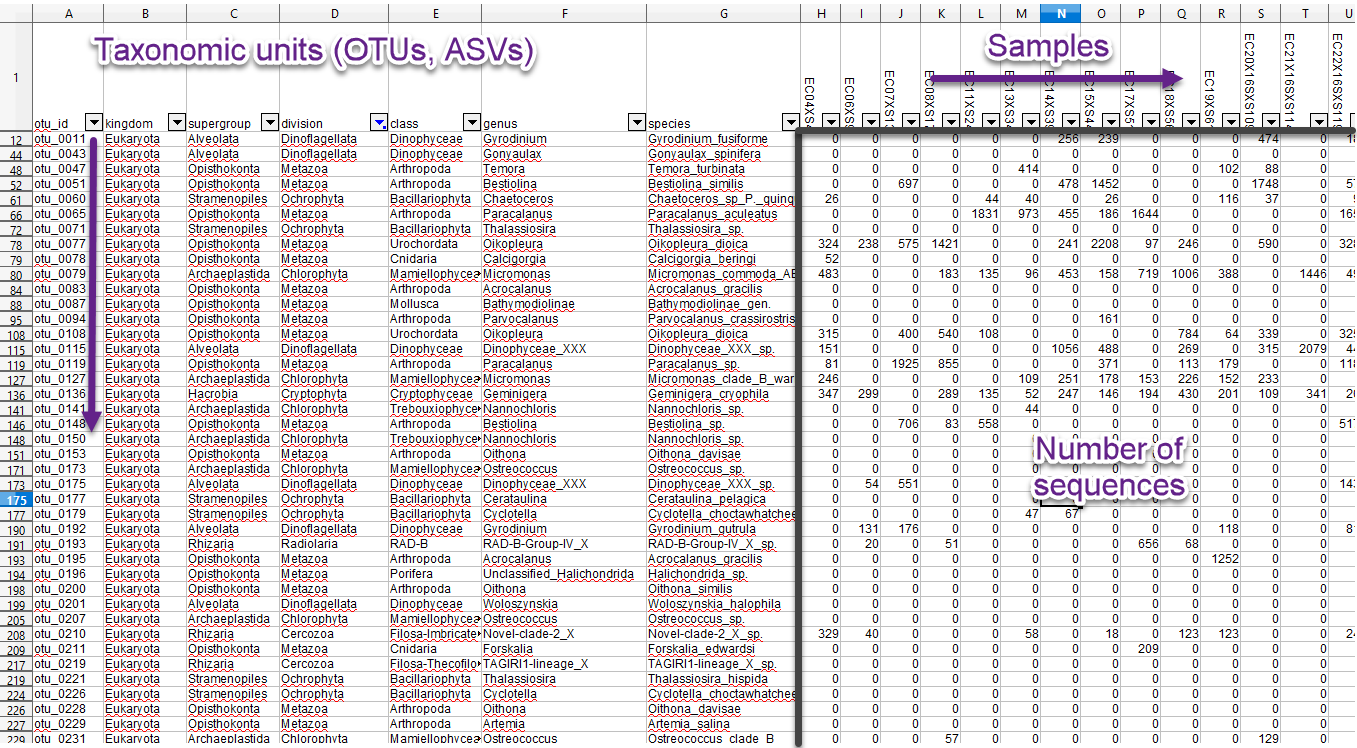
Data tables
Factors affecting protist communities
Substrate
- Water
- Ice
- Sediment
- Soil
- Microbiome
Ecosystem
- Oceanic
- Coastal
- Rivers
- Lakes
- Terrestrial
Size fraction
- Total (0.2 µm -> 100 µm)
- Pico (0.2 µm -> 2-3 µm)
- Nano (2-3 µm -> 20 µm)
- Micro (20 µm -> 100-200 µm)
- Meso (100 µm -> 1000 µm)
Factors affecting protist communities
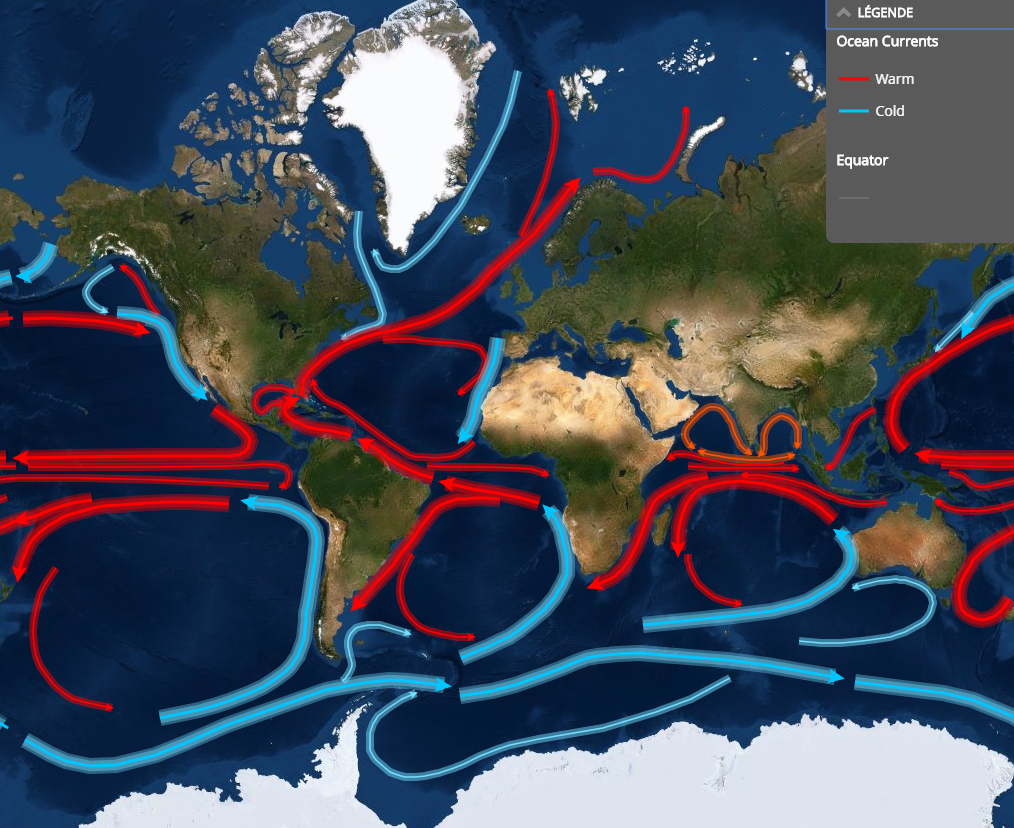
Environmental conditions
In oceanic waters:
- temperature
- salinity
- light
- nutrients
… which depend on:
- substrate (water vs.ice)
- latitude
- time of the year
- depth
- oceanic currents
- proximity of coast
Diversity
Microbial species in a sample
- species richness: total number of species
- species abundance: proportion of each species
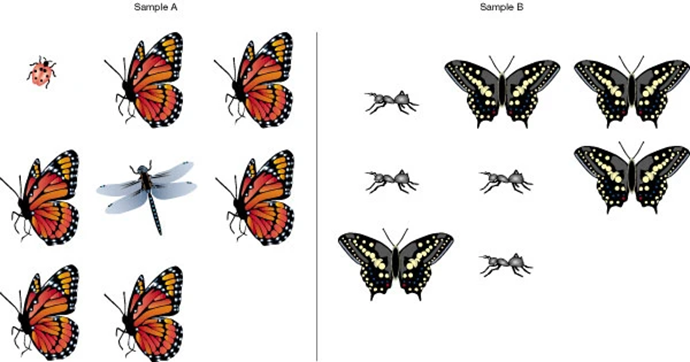
Richness vs. Evenness
Diversity
Alpha diversity - Diversity within a given sample
- Chao 1 is a non-parametric estimator of the number of species in a community.
- Shannon index1
\(H = - \sum_{i=1}^{S} p_i \cdot \log{p_i}\)
Where:
\(p_i\) = fraction of the entire population made up of species \(i\) (proportion of a species i relative to total number of species present)
\(S\) = numbers of species encountered
A high value of \(H\) would be a representative of a diverse and equally distributed community and lower values represent less diverse community. A value of 0 would represent a community with just one species.
Diversity
Alpha diversity - Effect of latitude
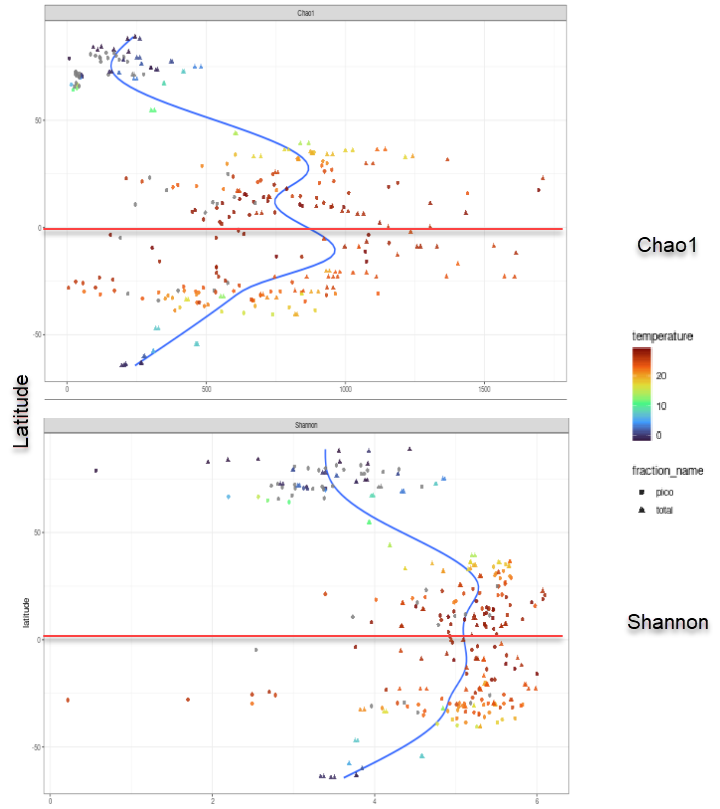
Diversity
Beta diversity - Compare diversity between samples
Compute distance between samples:
Bray-Curtis dissimilarity: use abundance information
- Varies between 0 and 1:
- 0 means the two samples have the same composition
- 1 means the two samples do not share any species
\(BC_{jk} = 1 - \frac{2\sum_{i=1}^{p}min(N_{ij},N_{ik})}{\sum_{i=1}^{p}(N_{ij} + N_{ik})}\)
where \(N_{ij}\) is the abundance of species \(i\) in sample \(j\) and \(p\) the total number of species
Jaccard similarity index
- Number of common species between samples divided by total number of species in the two samples \(J(A,B) = \frac{|A \cap B|}{|A \cup B|}\)
- Ordinate the samples
- NMDS: Non-Metric Multidimensional Scaling
Diversity
Beta diversity - Effect of depth on Stramenopiles communities
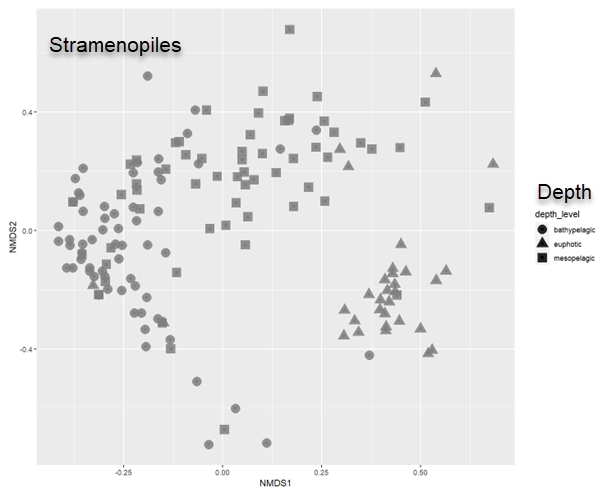
MetaPR2 - Datasets
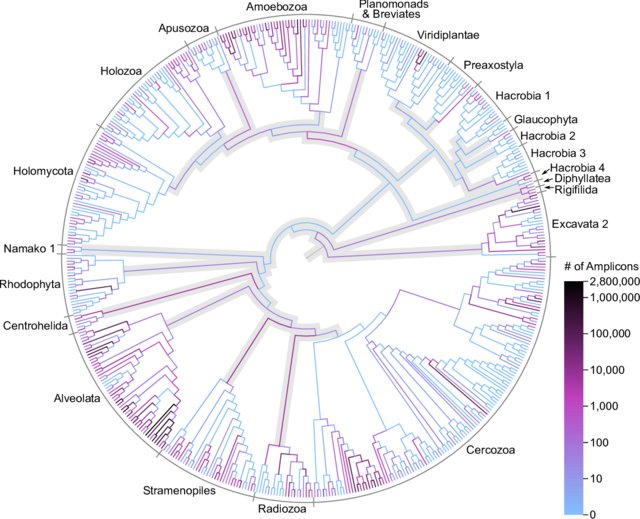
MetaPR2 - Taxonomy
Eight levels:
- Kingdom: Eukaryota
- Supergroup: Archaeplastida
- Division: Chlorophyta
- Class: Mamiellophyceae
- Order: Mamielliales
- Family: Bathycoccaceae
- Genus: Bathycococcus
- Species: B. prasinos
MetaPR2 - Visualization
MetaPR2 - In practice
Help
- Read in detail
Sample table
- dataset_name
- paper (can be useful to read)
- number of samples
- number of ASVs
- number of reads per sample (coverage)
Sample selection
- Major datasets: OSD, Tara, Malaspina
- By habitat: oceanic, coastal etc…
- Start by “marine global V4”
- Extend to other habitats/datasets
- V4 vs V9
- DNA vs. RNA
- Ecosystems
- Sustrate: water, ice, soil…
- Size fractions: total, pico…
- Depth level: surface, euphotic…
- Minimum ASV: will filter out rare ASVs (e.g. 1000)
- Selection can be saved (yaml file)
MetaPR2 - In practice
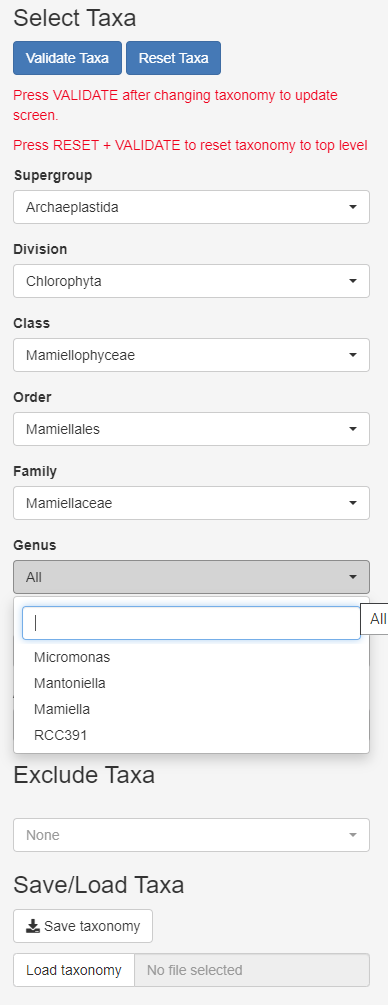
Taxonomy
- Can select several taxa within one level
- Press validate every time you need to refresh
- Can exclude taxa to remove fungi, metazoa…
- Can save taxonomy and reload taxonomy (yaml file)
MetaPR2 - In practice
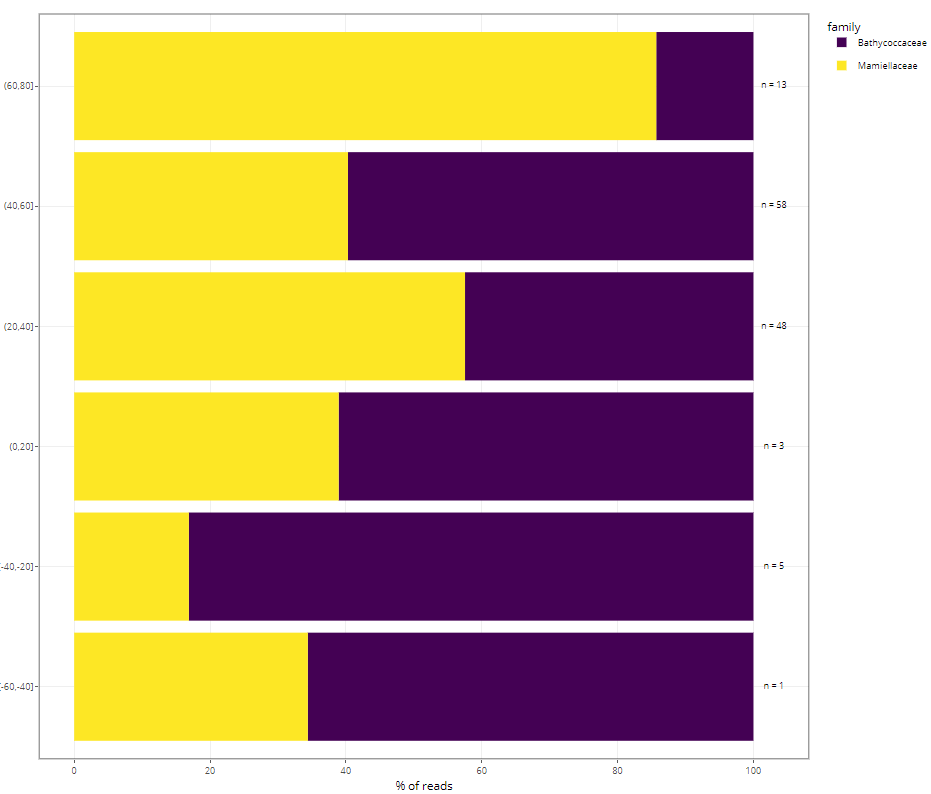
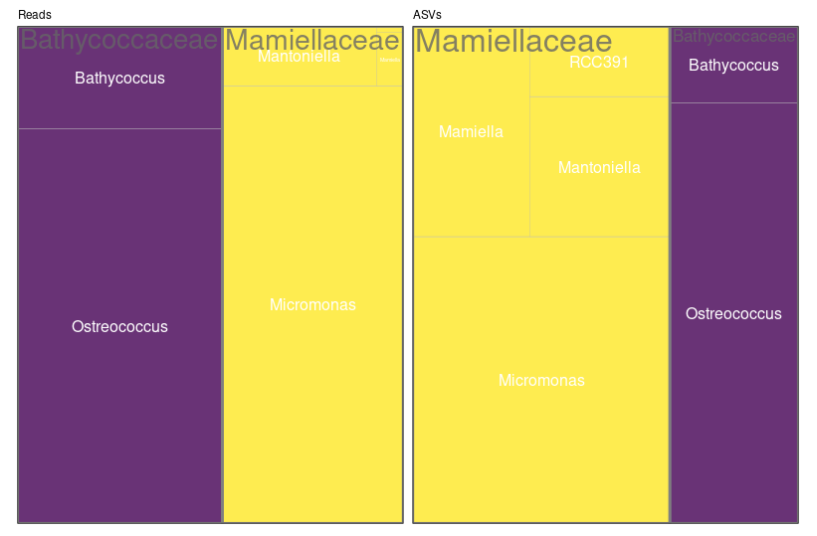
Treemaps
- Left panel: abundance (number of reads)
- Reads are “normalized” to 100
- Right panel: diversity (number of ASVs)
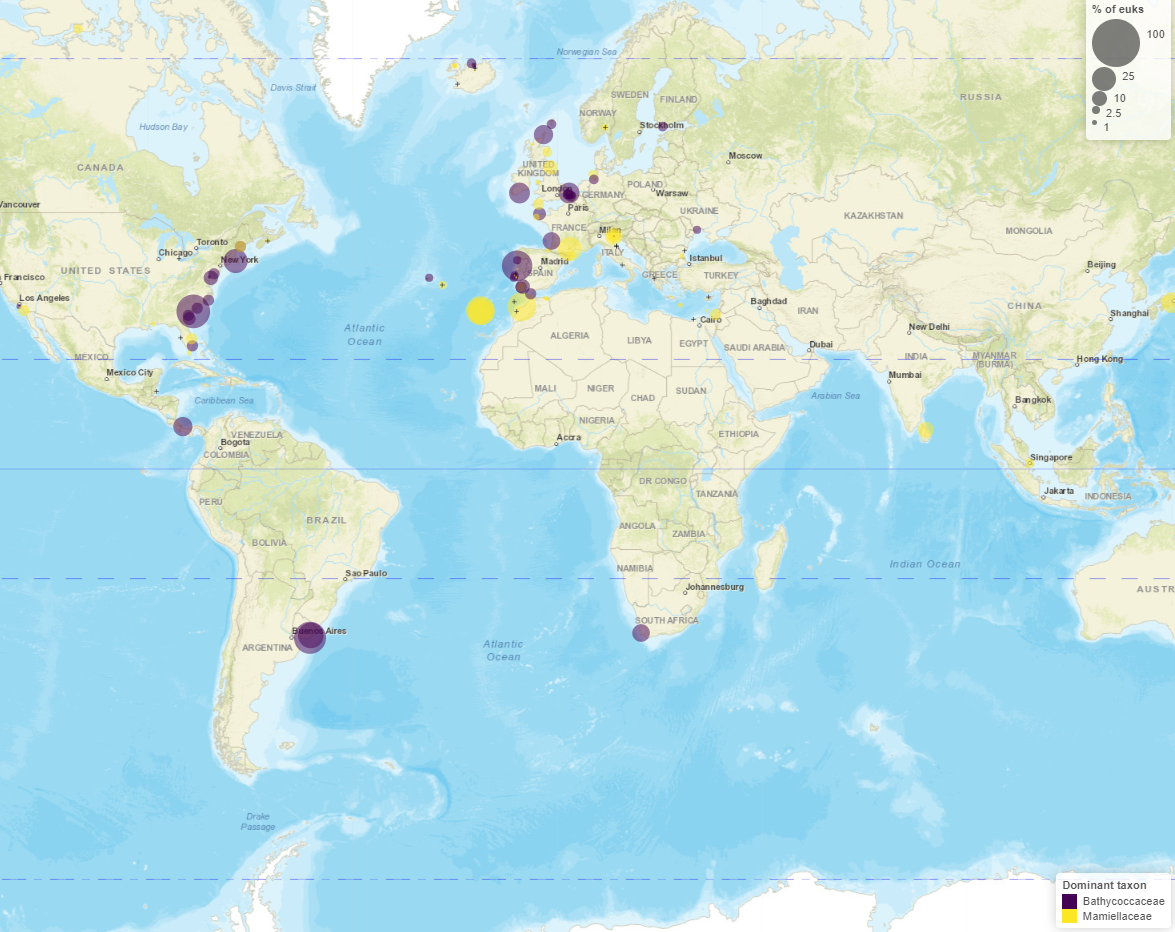
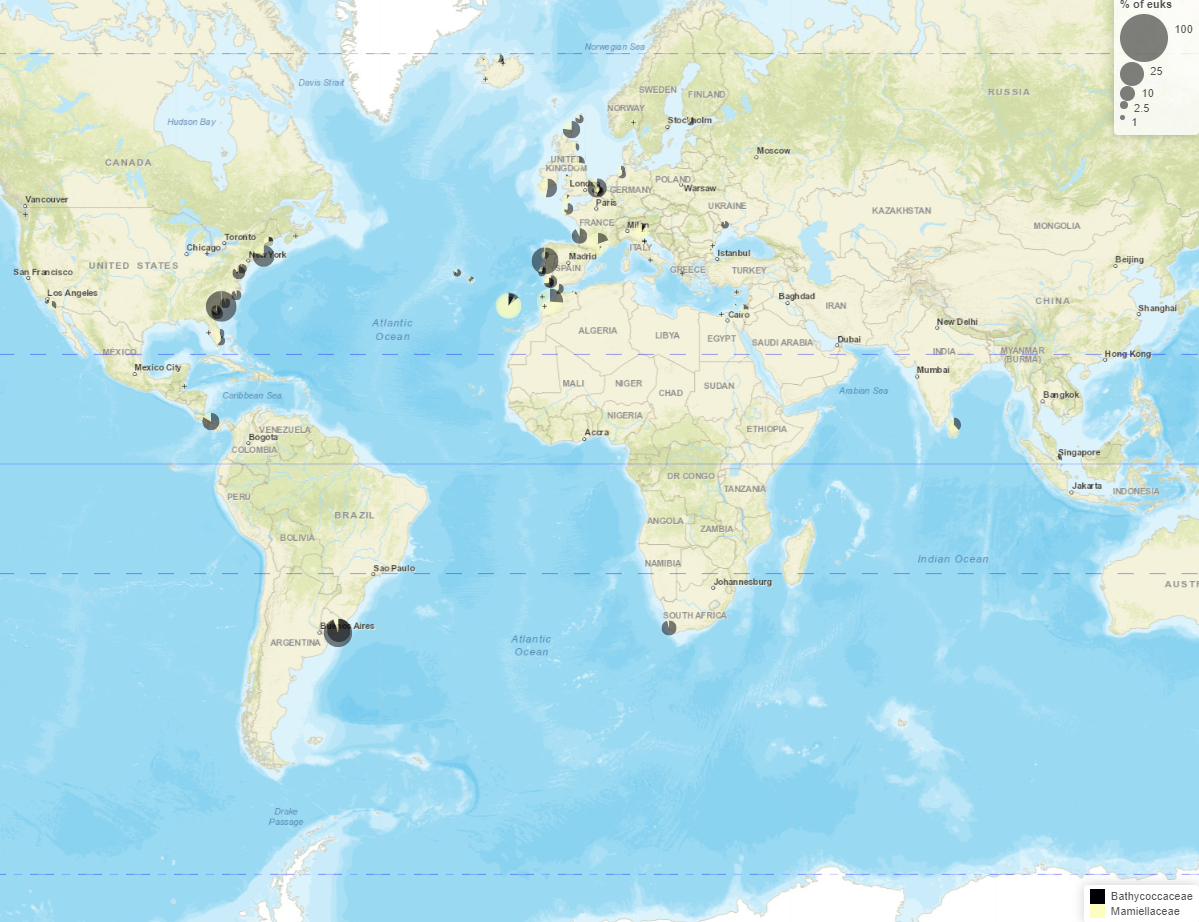
Maps
- Read information at top
- Taxo level
- Number of samples with/without taxa
- Crosses where taxa absent
- Map types
- Dominant
- Pie chart
- Circle scale
- Moving right increases size
Barplots
- taxonomy vs. function
- variables to use (but this depends on samples selected !)
- fraction name
- ecosystem
- substrate
- depth level
- DNA_RNA
- latitude
- temperature
- salinity
- year, month, day for time series
MetaPR2 - In practice
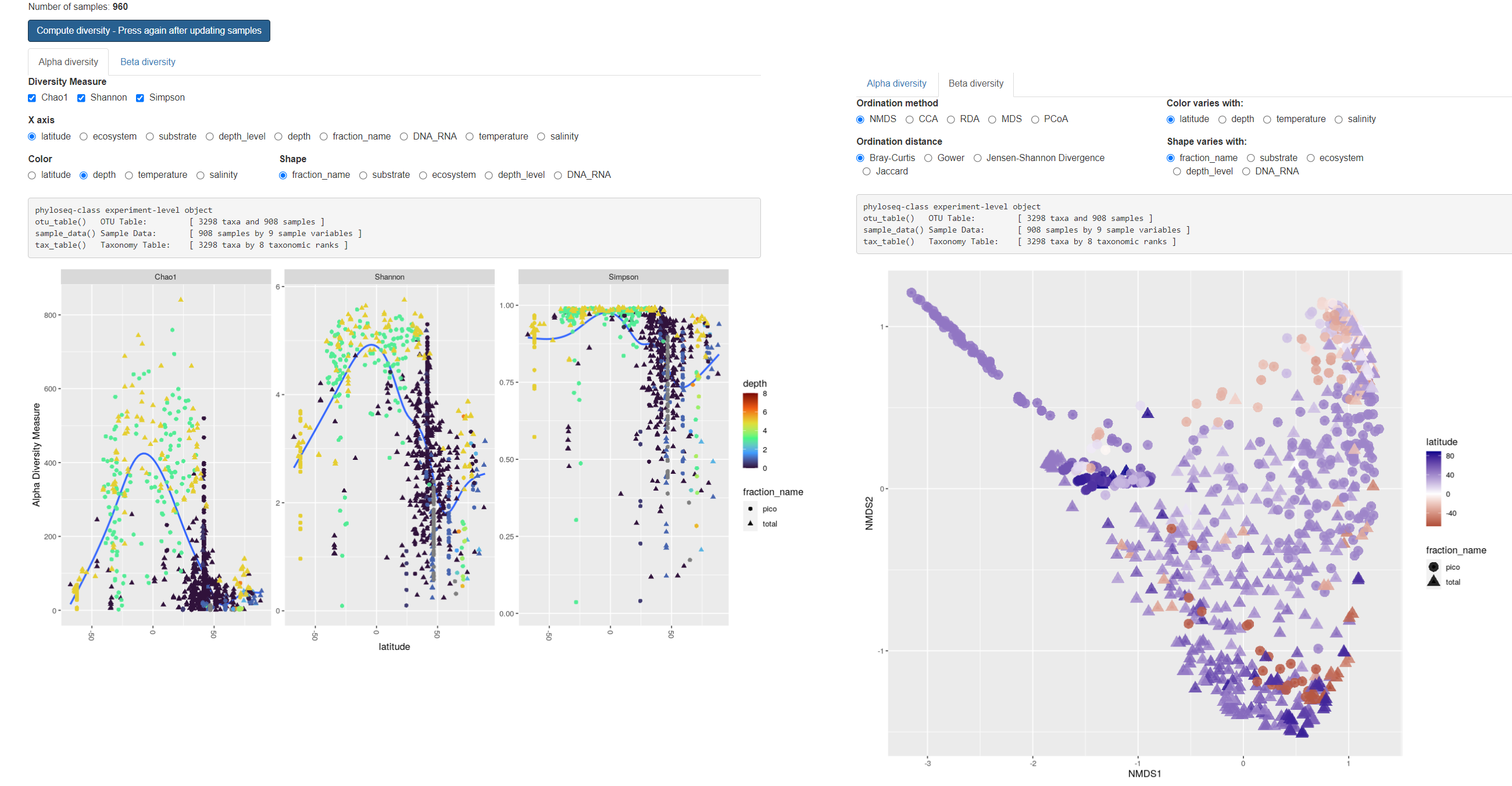
Diversity
- Hit “Compute…” after refreshing taxonomy
- Time proportional to N samples and taxa
- Information about
- Number of samples
- Number of taxa (ASVs)
Alpha diversity
- X: Chao1, Shannon, Simpson (compare)
- Discretize continuous Y
- Change Y (see barplots)
- Change shape
- Change color
Beta diversity
- Ordination method (difference ?)
- Ordination distance (Bray, Jaccard…)
- Change color and shape
MetaPR2 - In practice
Download
- Download
- datasets
- samples
- asv list with taxonomy
- asv sequences
Only for those with extensive experience with data processing.
Final presentation
Taxonomic groups
Green algae
- Prasinoderma
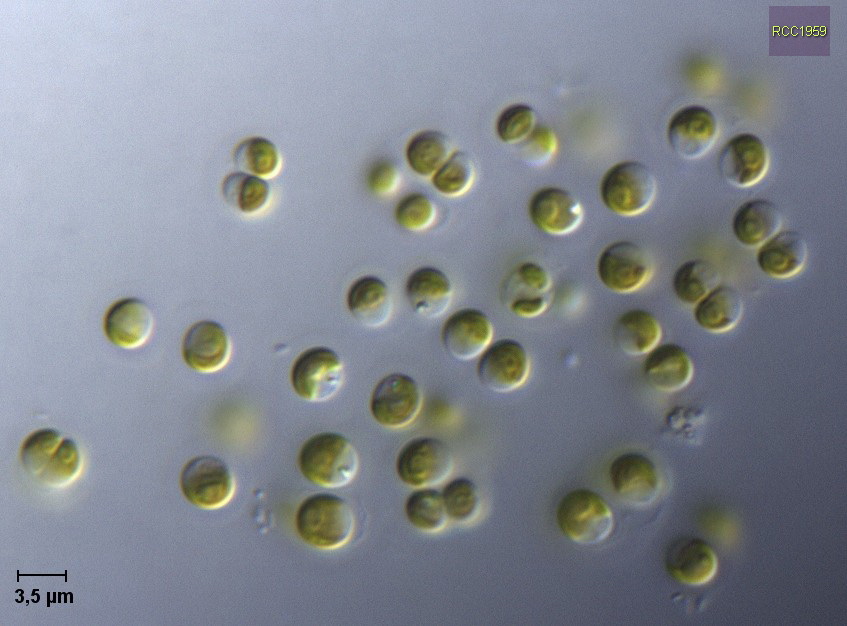
- Ostreococcus
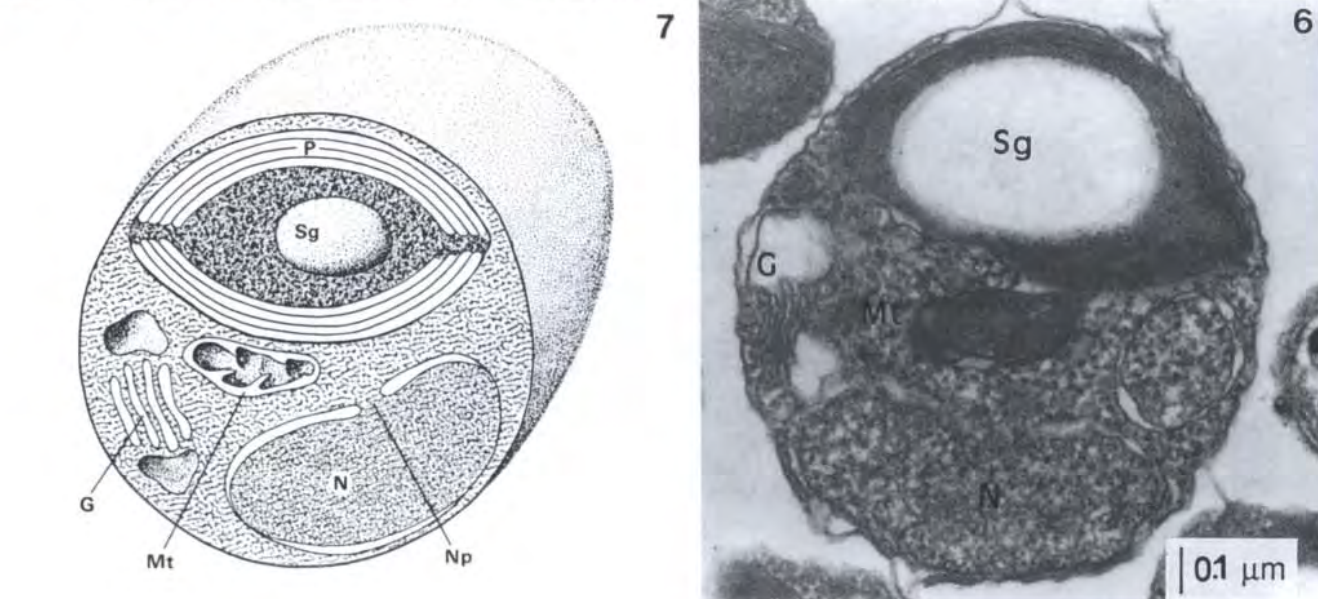
Ochrophyta (Stramenopiles)
- Pelagomonas, Aureococcus
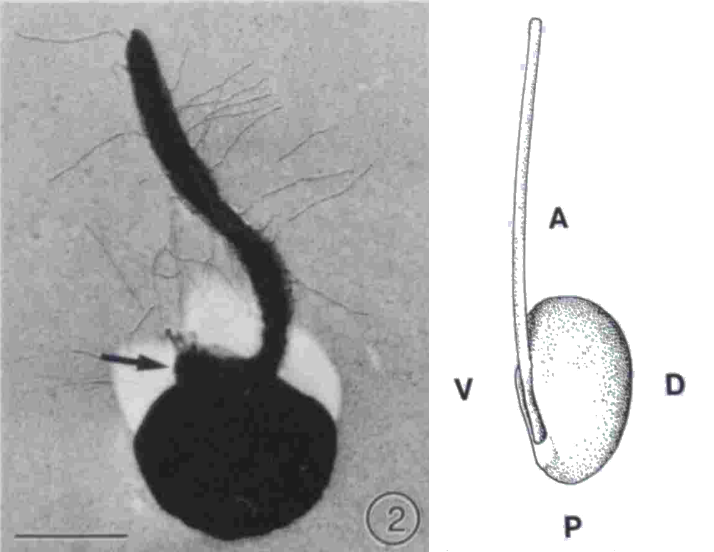
- Florenciella
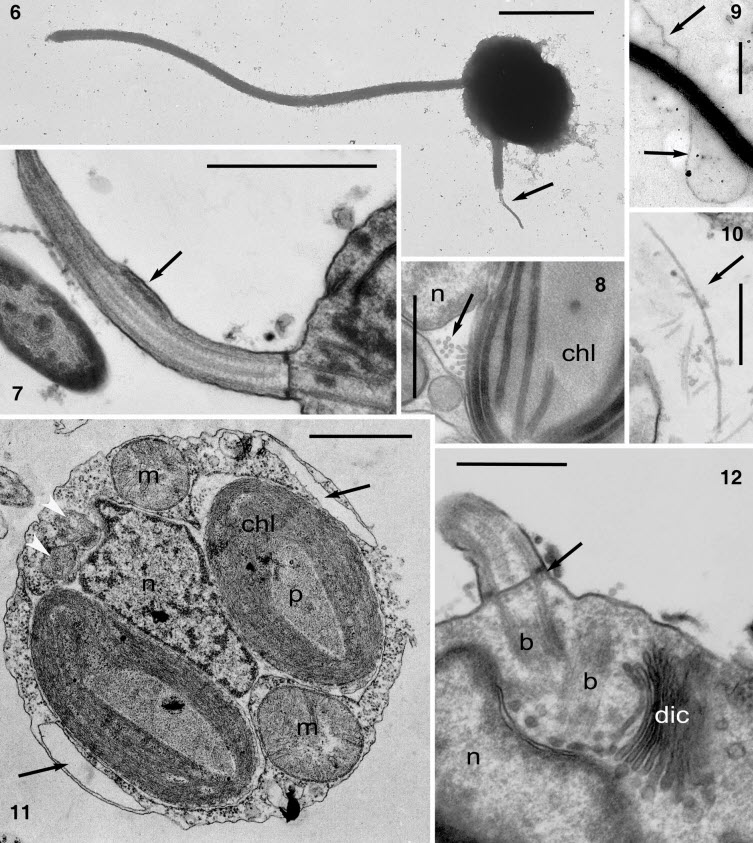
- Pinguiophyceae
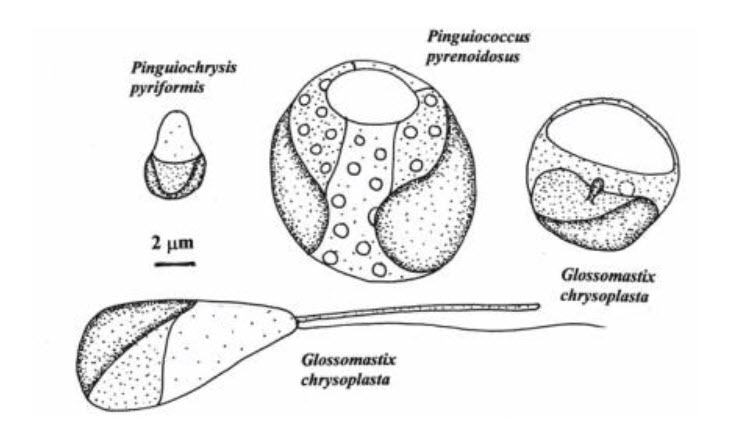
Final presentation
Taxonomic groups
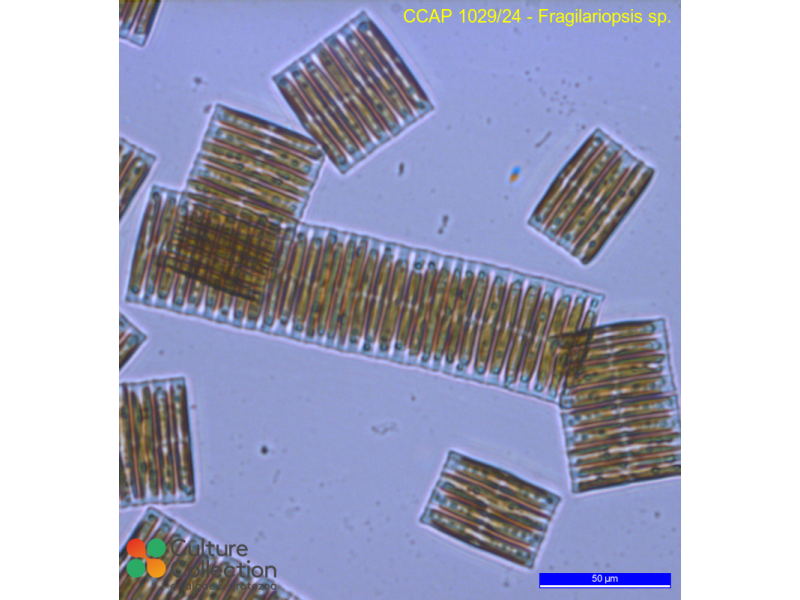
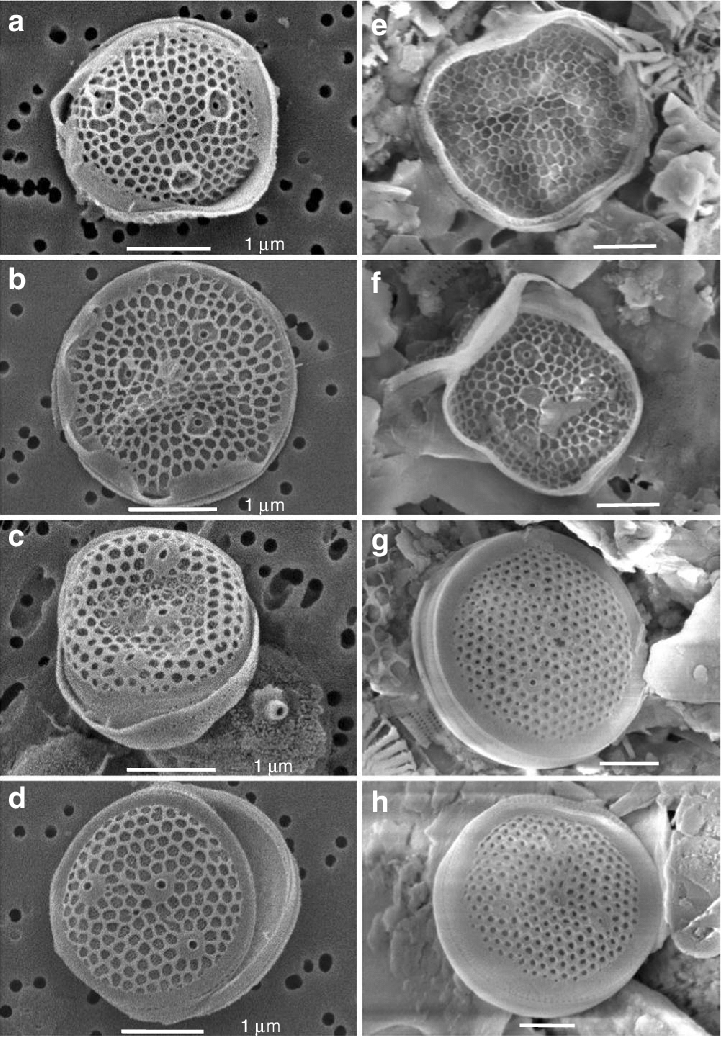
Diatoms
- Pseudo-nitzschia
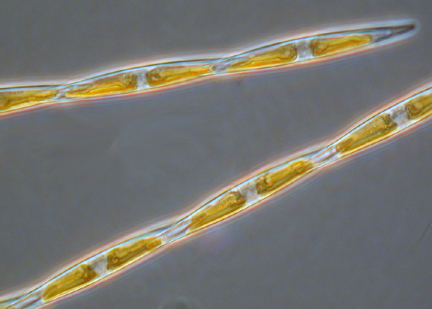
- Fragiliaropsis
- Minidiscus
- Rhizosolenia
Dinoflagellates
- Dinophysis
- Ceratium, Tripos

Final presentation
Key points
- Look for key papers on this group
- What are the dominant species?
- What is the microdiversity [diversity within dominant species (ASVs)]?
- What is distribution ?
- Substrate (water, ice…)
- Ecosystems (marine, freshwater, terrestrial)
- Size fraction
- Depth layers (euphotic zone vs. meso and bathypelagic)
- Latitudinal bands (polar, temperate, tropical)
- Coastal vs Pelagic
- Alpha diversity
- Beta diversity
Final presentation
In practice
- Each group will have max of 15’ to present their results. Your time will be cut after 15’.
- Don’t overload your presentation and run when talking. This will decrease the clarity of your presentation.
- Share equally time between group members.
- Introduce very briefly the main biological characteristics and ecological importance of your taxonomic group.
- Explain which hypotheses/questions your group were interested in.
- Explain the results you have observed. Focus on main points.
- Each group will have 5’ to answer questions.
Final presentation
Evaluation
- Profs, TAs and PhD’s students will be judging your presentation (Only Profs will grade!):
- Grade scale: 0 = unacceptable; 1 = poor; 2 = fair; 3 = good; 4 = outstanding
Criteria
- Quality of presentation
- Slides (font size, amount on slide, legible and clear, references, no errors, etc).
- Organization of presentation (outline, logical sequence, good transitions, easy to follow, etc).
- Quality of oral presentation (well paced, projected voice, face audience, eye contact, confident, etc).
- Did the group keep the audience interested? (show enthusiasm, command attention, did you learn something new?)
- Was the presentation within the 15 minutes in length?
- Content of presentation
- Was the presentation well structured ?
- Did the group show an overall understanding of the topic? (background, objectives and significance thoroughly explained?).
- Did the presentation cited the relevant material from the litterature?
- Did the group answered questions accurately? Did the group possess good understanding of topic based on answers?