Microbes on Earth
R session 03 - Data viz
Daniel Vaulot
2021-10-12
1 / 40
Outline
- Graph types
- Grammar of graphics
- Playing with ggplot2
- Multiple graphs
- ggplot2 syntax
2 / 40
Installation and Resources
Packages
- ggplot2
- patchwork
Download
- R-session-03.zip
Reading assignement
Resources

3 / 40
Data vizualization
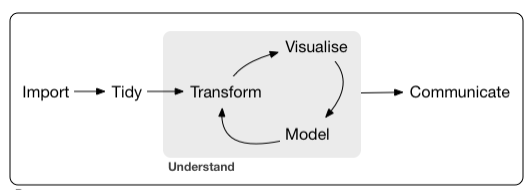
4 / 40
Data vizualization

Graph purposes
4 / 40
Data vizualization

Graph purposes
- Analysis graphs
- design to see patterns, trends
- aid the process of data description
- interpretation
4 / 40
Data vizualization

Graph purposes
- Analysis graphs
- design to see patterns, trends
- aid the process of data description
- interpretation
- Presentation graphs
- design to attract attention
- make a point
- illustrate a conclusion
Source: Michael Friendly - http://datavis.ca/courses/RGraphics/
4 / 40
Graph types
Jitter
- Two variables numerical
5 / 40
Graph types
Jitter
- Two variables numerical
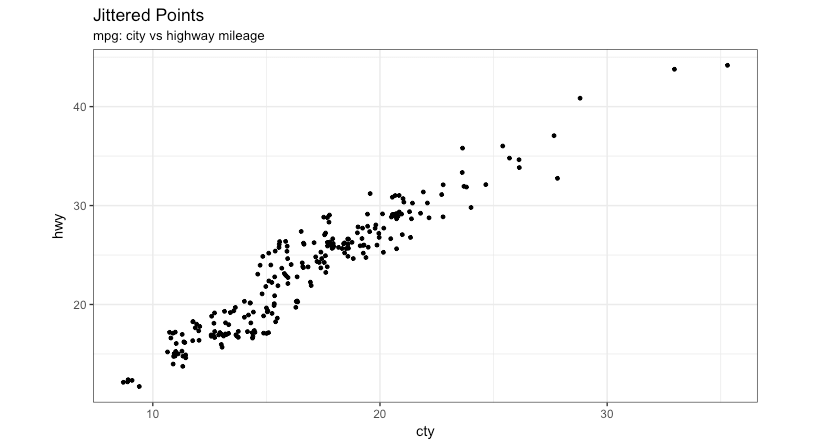
5 / 40
Graph types
Bubble
- Two variables numerical
- Add another variable numerical
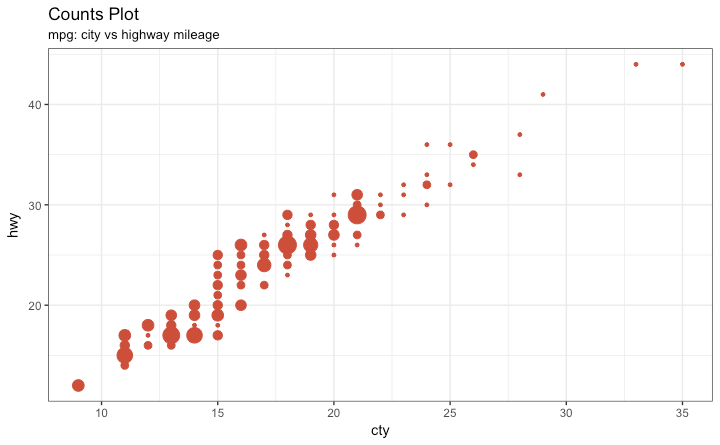
6 / 40
Graph types
Bargraphs
- One variable categorical
- One variable numerical
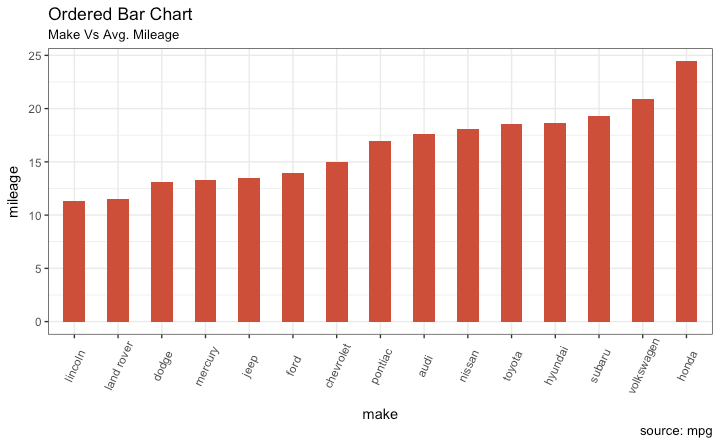
7 / 40
Graph types
Bargraphs
- Rotate
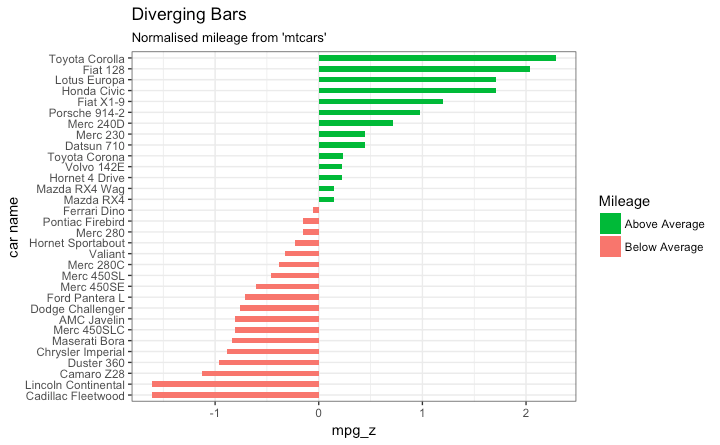
8 / 40
Graph types
Bargraphs
- Two variable categorical
- One variable numerical
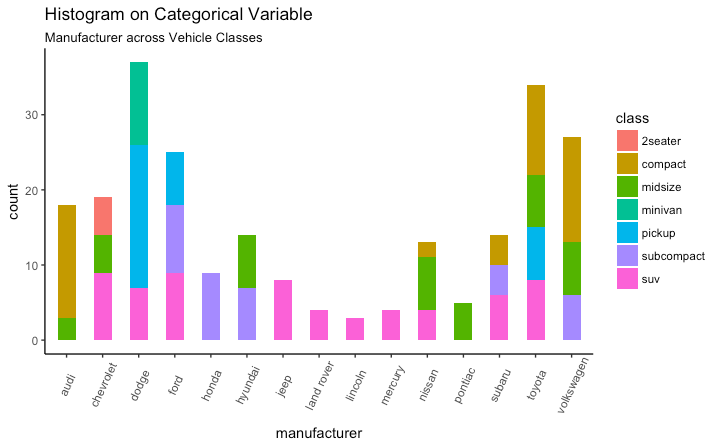
9 / 40
Graph types
Boxplots
- One variable categorical
- One variable numerical but with many values
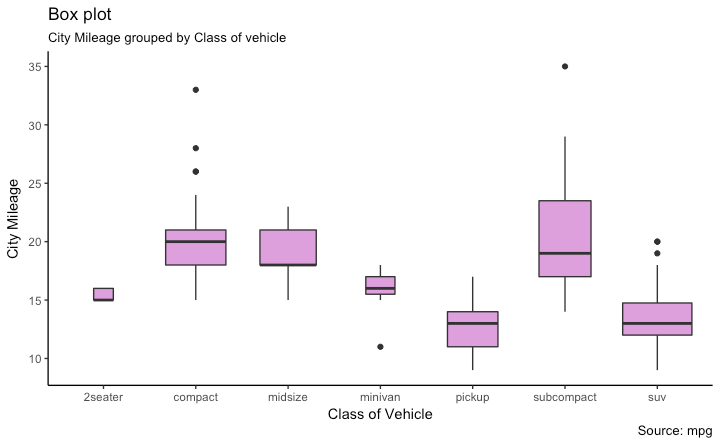
10 / 40
Graph types
Treemaps
- One variable categorical
- One variable numerical
- Much better than pie charts
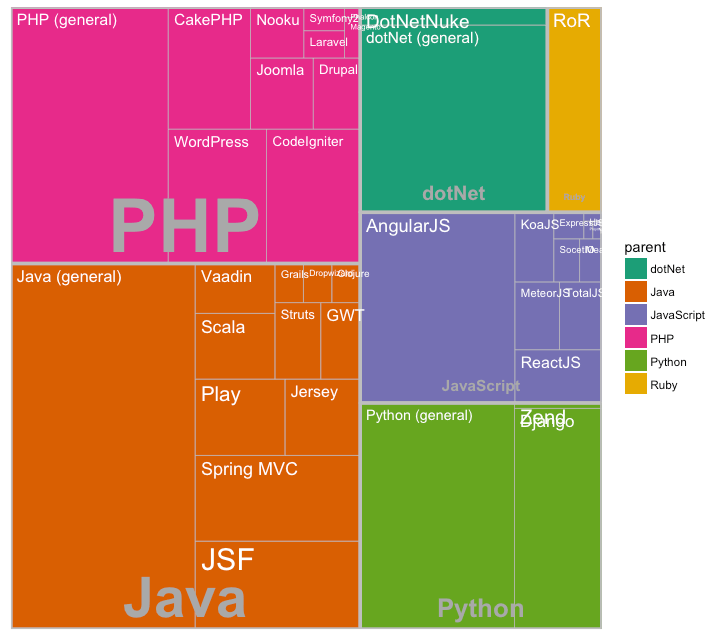
11 / 40
Graph types
Many...
- Choose as a function of what you want to analyze or the story you want to tell
- https://www.r-graph-gallery.com/all-graphs/
12 / 40
ggplot2

@allison_horst
13 / 40
Initialize
Load necessary libraries
library("readxl") # Import the data from Excel filelibrary("dplyr") # filter and reformat data frameslibrary("ggplot2") # graphics14 / 40
Initialize
Read the data
samples <- readxl::read_excel("data/CARBOM data.xlsx", sheet = "Samples_boat") %>% tidyr::fill(station)| sample number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 |
| 121 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 |
| 122 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 |
| 126 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 |
| 127 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 |
15 / 40
ggplot2
A simple plot
- Choose the data set
- Choose the geometric representation
- Choose the aesthetics : x,y, color, shape etc...
ggplot(data=samples) + geom_point(mapping = aes(x=phosphates, y=nitrates))- All functions are from ggplot2 package unless specified
16 / 40
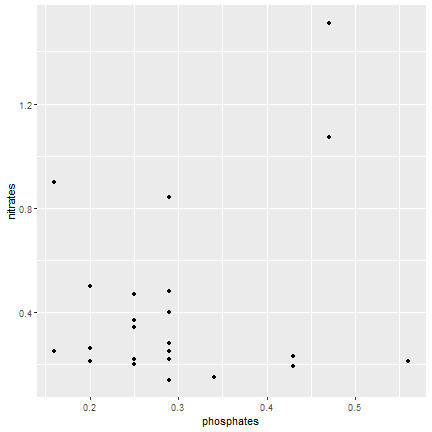
ggplot2
A simple plot
- Choose the data set
- Choose the geometric representation
- Choose the aesthetics : x,y, color, shape etc...
ggplot(data=samples) + geom_point(mapping = aes(x=phosphates, y=nitrates))- All functions are from ggplot2 package unless specified
16 / 40
ggplot2
The grammar of graphics
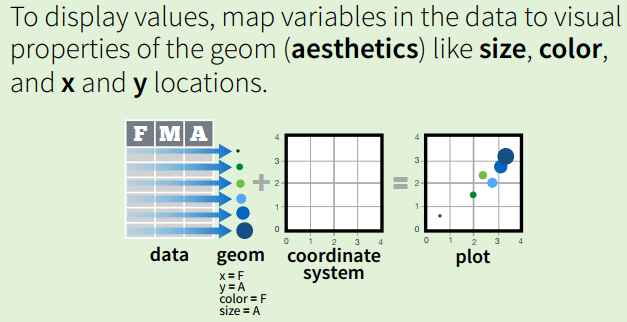
Every graph can be described as a combination of independent building blocks:
- data: a data frame: quantitative, categorical; local or data base query
- aesthetic mapping of variables into visual properties: size, color, x, y
- geometric objects (“geom”): points, lines, areas, arrows, …
- coordinate system (“coord”): Cartesian, log, polar, map
17 / 40
ggplot2
Syntax
ggplot(data=samples) + geom_point(mapping = aes(x=phosphates, y=nitrates))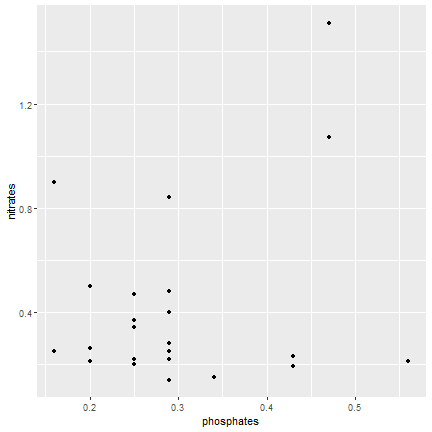
18 / 40
ggplot2
Alternatively
ggplot(data=samples, mapping = aes(x=phosphates, y=nitrates)) + geom_point()- If different geometries origniate from different datasets or have different mapping the datasets or the mapping must be called inside the geom function.
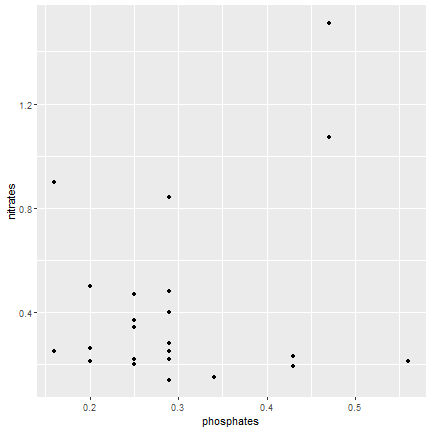
19 / 40
ggplot2
Alternatively
ggplot(samples, aes(x=phosphates, y=nitrates)) + geom_point()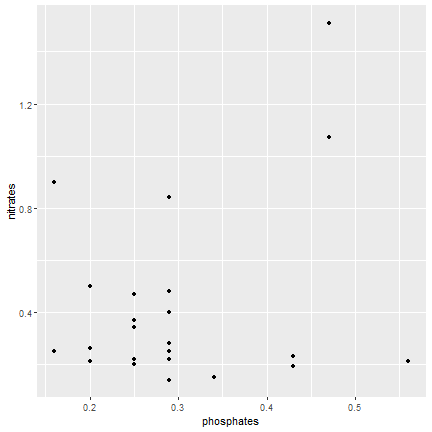
20 / 40
ggplot2
Make dot size bigger
ggplot(samples, aes(x=phosphates, y=nitrates))
21 / 40
ggplot2
Make dot size bigger
ggplot(samples, aes(x=phosphates, y=nitrates)) + geom_point(size=5)- Add: size=5 outside of the aesthetics function
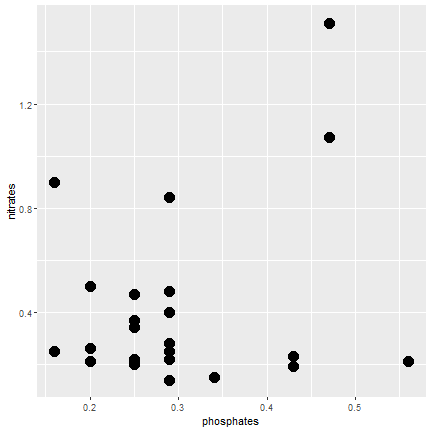
22 / 40
ggplot2
Color according to depth level (discrete)
ggplot(samples, aes(x=phosphates, y=nitrates, color=level)) + geom_point(size=5)- The mapping aesthetics must be an argument of the aes function
- geom_point(color=level, size=5) will generate an error...
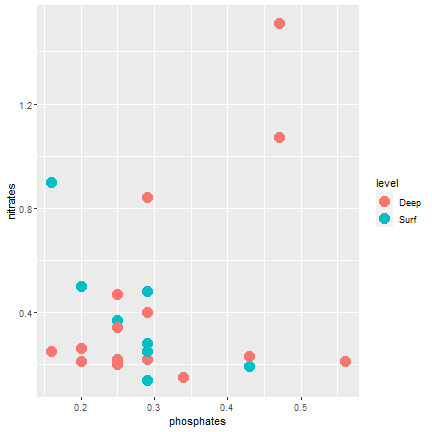
23 / 40
ggplot2
Color according to depth (continuous)
ggplot(samples, aes(x=phosphates, y=nitrates, color=depth)) + geom_point(size=5)- Add: color=depth
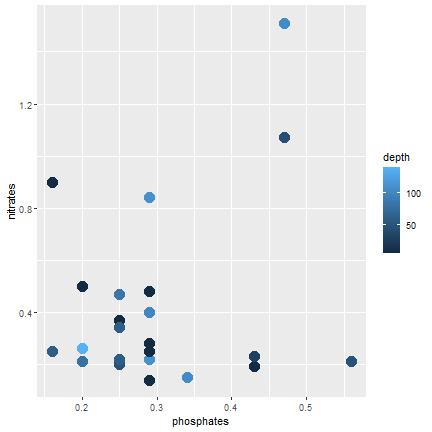
24 / 40
ggplot2
Symbol according to transect (continuous)
ggplot(samples, aes(x=phosphates, y=nitrates, color=depth, shape=transect)) + geom_point(size=5)- Add: shape=transect
Error: A continuous variable can not be mapped to shape
25 / 40
ggplot2
Symbol according to transect (continuous)
ggplot(samples, aes(x=phosphates, y=nitrates, color=depth, shape=as.character(transect))) + geom_point(size=5)- Add: shape=as.character(transect)
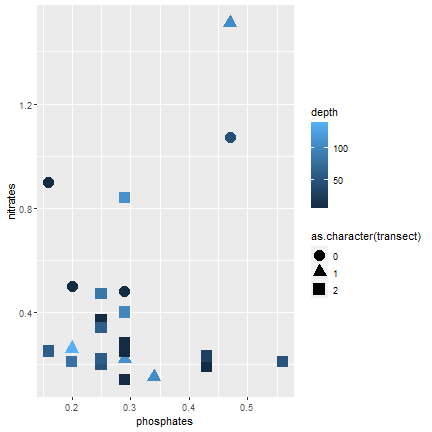
26 / 40
ggplot2
Panels depending on one variable
ggplot(samples, aes(x=phosphates, y=nitrates)) + geom_point() + facet_wrap(~ level)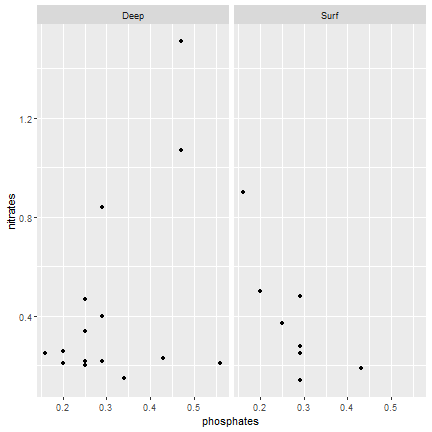
27 / 40
ggplot2
Adding a regression line
ggplot(samples, aes(x=phosphates, y=nitrates, color=level)) + geom_point(size=5) + geom_smooth(mapping = aes(x=phosphates, y=nitrates), method="lm")- Add: geom_smooth()
- You can choose the type of smoothing "lm" is for linear model
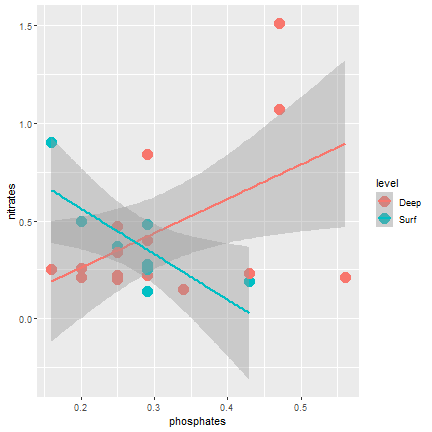
28 / 40
ggplot2
Adding a regression line
ggplot(samples, aes(x=phosphates, y=nitrates)) + geom_point(aes(color=level), size=5) + geom_smooth(mapping = aes(x=phosphates, y=nitrates), method="lm")- If the mapping is in the ggplot function is for all the geom....
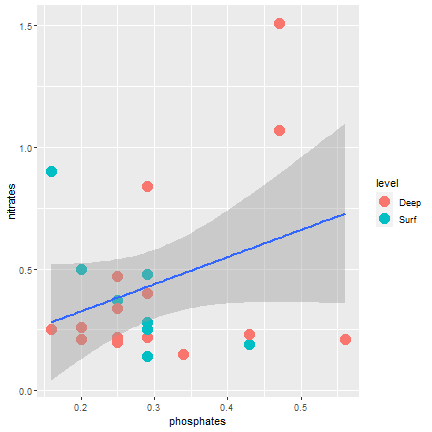
29 / 40
ggplot2
Finalizing the graph
ggplot(samples) + geom_point(mapping = aes(x=phosphates, y=nitrates, color=level), size=5) + geom_smooth(mapping = aes(x=phosphates, y=nitrates), method="lm") + xlab("Phosphates") + ylab("Nitrates") + ggtitle("CARBOM cruise")- Add: geom_smooth()
- You can choose the type of smoothing "lm" is for linear model
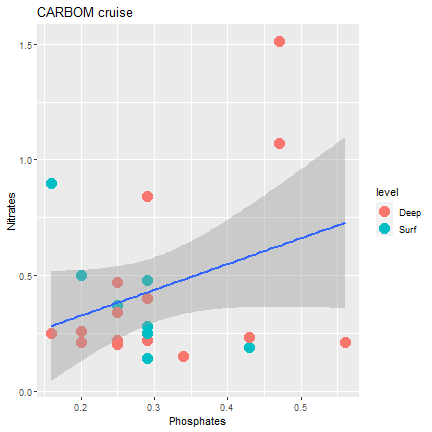
30 / 40
ggplot2 syntax
Anatomy of a plot
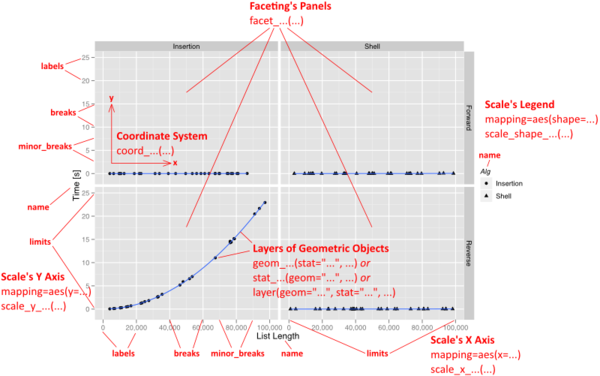
31 / 40
ggplot2 syntax
Geometries
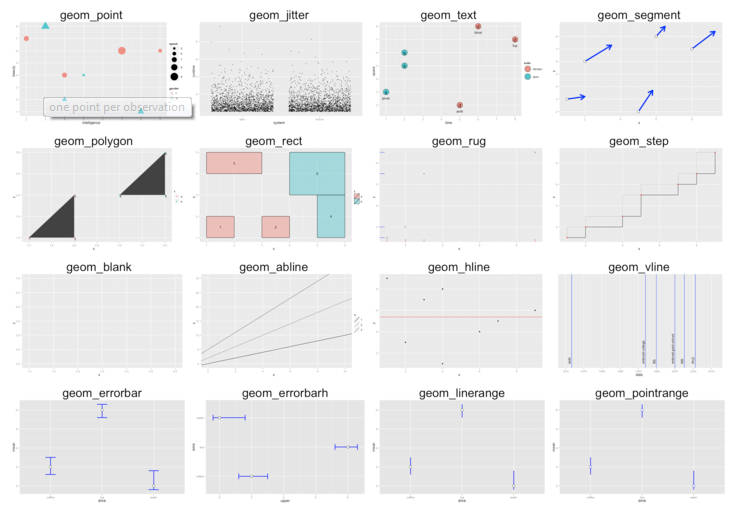
32 / 40
ggplot2 syntax
Continuous x and y

33 / 40
ggplot2 syntax
Plotting error

34 / 40
ggplot2 syntax
Discrete x - Continuous y

35 / 40
ggplot2 syntax
Continuous x
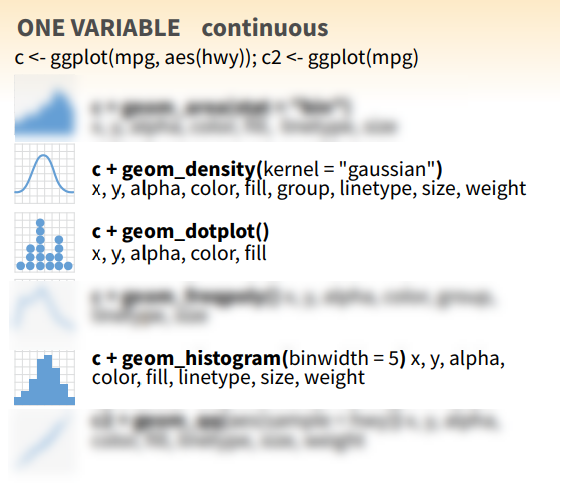
36 / 40
ggplot2 syntax
Modifying axis and scales
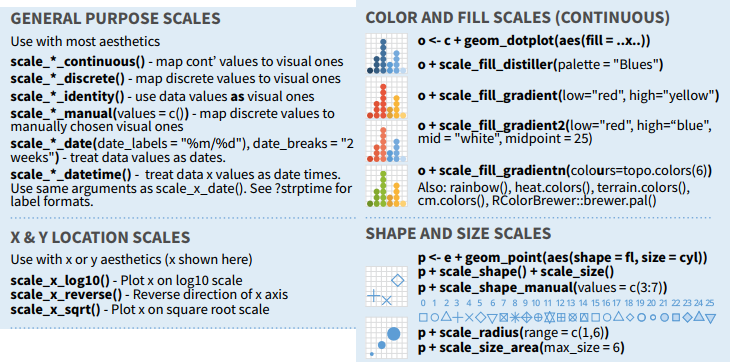
37 / 40
ggplot2 syntax
Palettes
Package tmaptools : https://github.com/mtennekes/tmaptools
- Function :
palette_explorer()
- Function :
Package paletteer : https://github.com/EmilHvitfeldt/paletteer
- More than 1000 palettes
38 / 40
ggplot2 syntax
Palettes
- Use color blind friendly palettes
- viridis (e.g. scale_colour_viridis_c)
39 / 40
Recap
- Conceptualize your graph before coding
40 / 40
Recap
- Conceptualize your graph before coding
- Decide what element is fixed and what varies
40 / 40
Recap
- Conceptualize your graph before coding
- Decide what element is fixed and what varies
- It takes time to get what you want...
40 / 40
Recap
- Conceptualize your graph before coding
- Decide what element is fixed and what varies
- It takes time to get what you want...
- Exploratory vs. final
40 / 40