R course
Daniel Vaulot
2023-01-26



Metabarcode analysis - Phyloseq
R - Phyloseq
- Introduction to the data
- Read the data
- Format and save as Phyloseq objects
- Filter data
- Bar graphs
- Alpha diversity
- Beta diversity
Phyloseq R library
See in particular tutorials for
Get ASVs, read abundance and metadata all together
Filter and regroup the data
Bar plots, Alpha and Beta diversity computations
Data
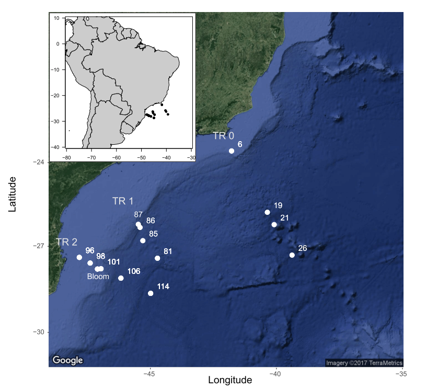
This tutorial uses a reduced metabarcoding dataset obtained by C. Ribeiro and A. Lopes dos Santos. This dataset originates from the CARBOM cruise in 2013 off Brazil and corresponds to the 18S V4 region amplified on flow cytometry sorted samples (see pptx file for details) and sequenced on an Illumina run 2*250 bp analyzed with mothur.
Reference
- Gérikas Ribeiro, C., Dos Santos, A.L., Marie, D., Brandini, F.P. & Vaulot, D. 2018. Small eukaryotic phytoplankton communities in tropical waters off Brazil are dominated by symbioses between Haptophyta and nitrogen-fixing cyanobacteria. ISME Journal. 12:1360–74.
Libraries
Import and format data
Three tables are needed
- OTU
- Taxonomy
- Samples
They are read from a single Excel file where each sheet contains one of the tables
OTU
- rows = OTUs or ASVs
- cols = samples
- cells = number of reads

Table OTU - OTU abundance
Taxonomy
- rows = OTUs or ASVs
- cols = taxonomy ranks
- cells = taxon name
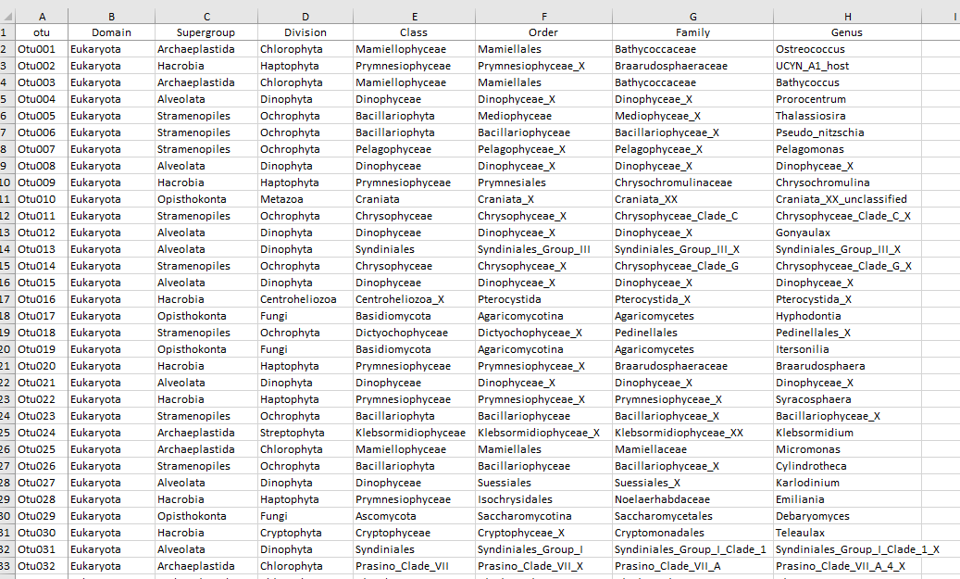
Table Taxo - OTU taxonomy
Samples
- rows = samples
- cols = metadata type
- cells = metadata values
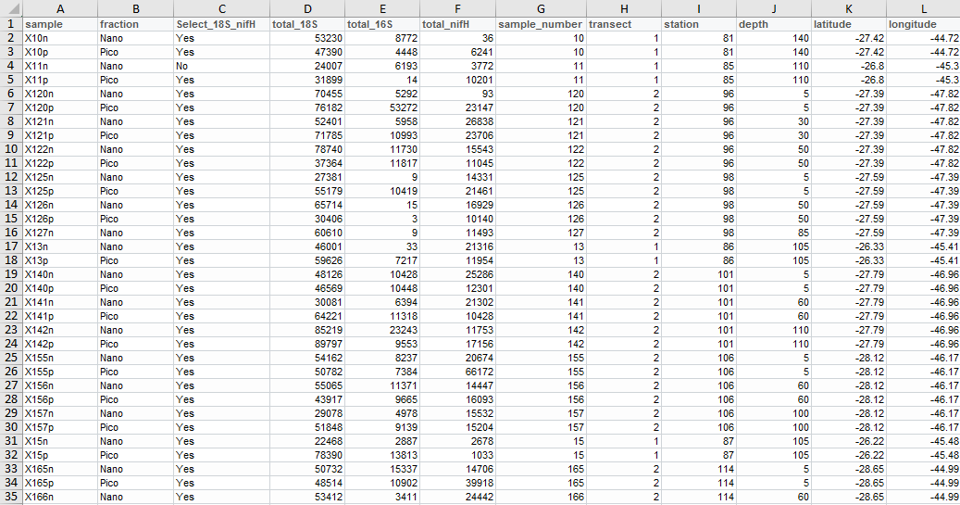
Table Samples
Read Excel files
Phyloseq objects need to have row.names
- define the row names from the otu column
- Idem for the two other matrixes
Transform to matrixes
Sample table can be left as data frame
Transform to phyloseq objects
OTU = otu_table(otu_mat, taxa_are_rows = TRUE)
TAX = tax_table(tax_mat)
samples = sample_data(samples_df)
carbom <- phyloseq(OTU, TAX, samples)
carbomphyloseq-class experiment-level object
otu_table() OTU Table: [ 287 taxa and 55 samples ]
sample_data() Sample Data: [ 55 samples by 27 sample variables ]
tax_table() Taxonomy Table: [ 287 taxa by 7 taxonomic ranks ]Check data
[1] "X10n" "X10p" "X11n" "X11p" "X120n" "X120p" "X121n" "X121p"
[9] "X122n" "X122p" "X125n" "X125p" "X126n" "X126p" "X127n" "X13n"
[17] "X13p" "X140n" "X140p" "X141n" "X141p" "X142n" "X142p" "X155n"
[25] "X155p" "X156n" "X156p" "X157n" "X157p" "X15n" "X15p" "X165n"
[33] "X165p" "X166n" "X166p" "X167n" "X167p" "X1n" "X1p" "X2n"
[41] "X2p" "X3n" "X3p" "X5n" "X5p" "X7n" "X7p" "X9n"
[49] "X9p" "tri01n" "tri01p" "tri02n" "tri02p" "tri03n" "tri03p"[1] "Domain" "Supergroup" "Division" "Class" "Order"
[6] "Family" "Genus" [1] "fraction" "Select_18S_nifH" "total_18S"
[4] "total_16S" "total_nifH" "sample_number"
[7] "transect" "station" "depth"
[10] "latitude" "longitude" "picoeuks"
[13] "nanoeuks" "bottom_depth" "level"
[16] "transect_distance" "date" "time"
[19] "phosphates" "silicates" "ammonia"
[22] "nitrates" "nitrites" "temperature"
[25] "fluorescence" "salinity" "sample_label" Filter and normalize data
Keep only samples to be analyzed
Keep only photosynthetic taxa
carbom <- subset_taxa(carbom, Division %in% c("Chlorophyta", "Dinophyta", "Cryptophyta",
"Haptophyta", "Ochrophyta", "Cercozoa"))
carbom <- subset_taxa(carbom, !(Class %in% c("Syndiniales", "Sarcomonadea")))
carbomphyloseq-class experiment-level object
otu_table() OTU Table: [ 205 taxa and 54 samples ]
sample_data() Sample Data: [ 54 samples by 27 sample variables ]
tax_table() Taxonomy Table: [ 205 taxa by 7 taxonomic ranks ]Normalize
Normalize number of reads in each sample using median number of reads.
The number of reads used for normalization is 44903.
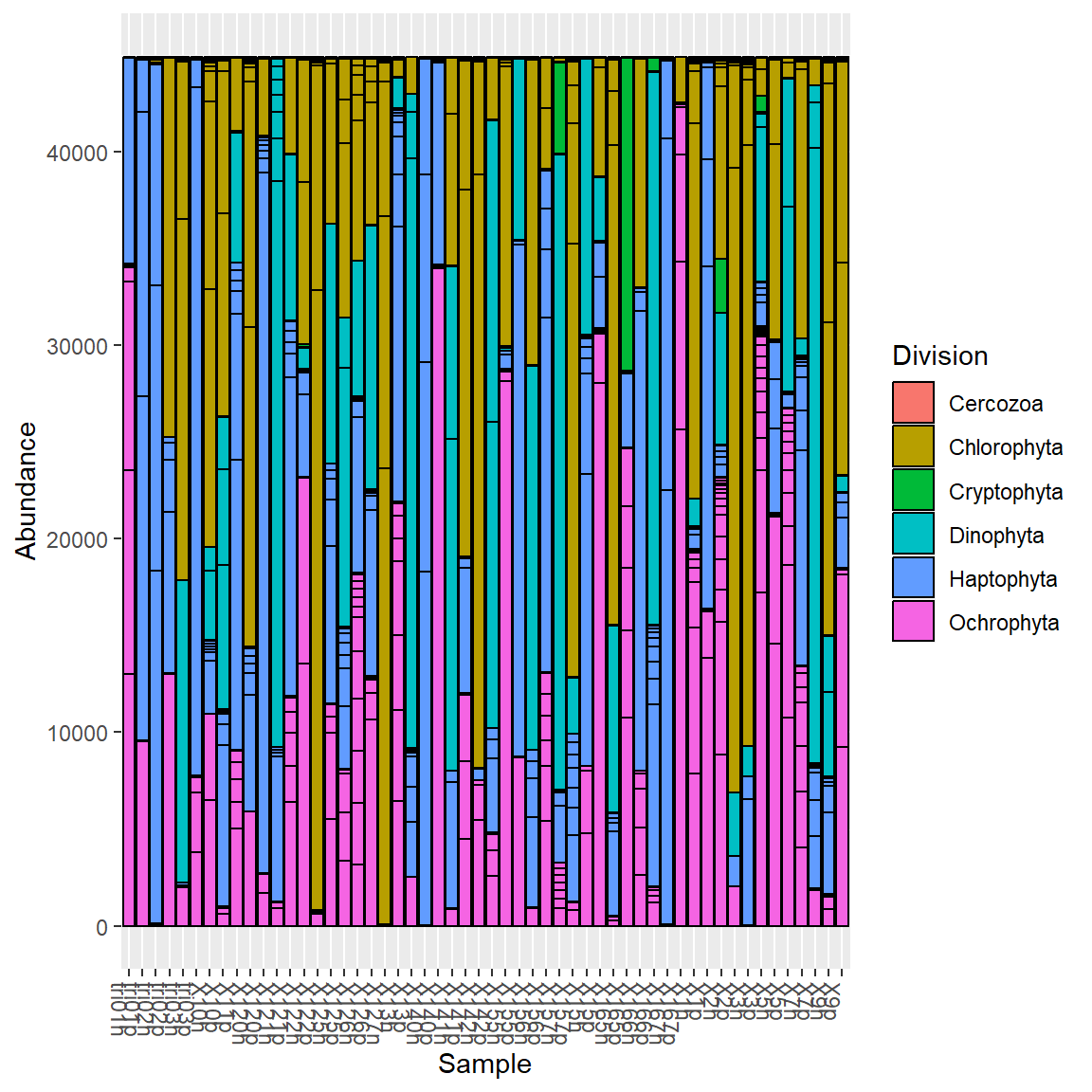
Bar graphs
Basic bar graph based on Division
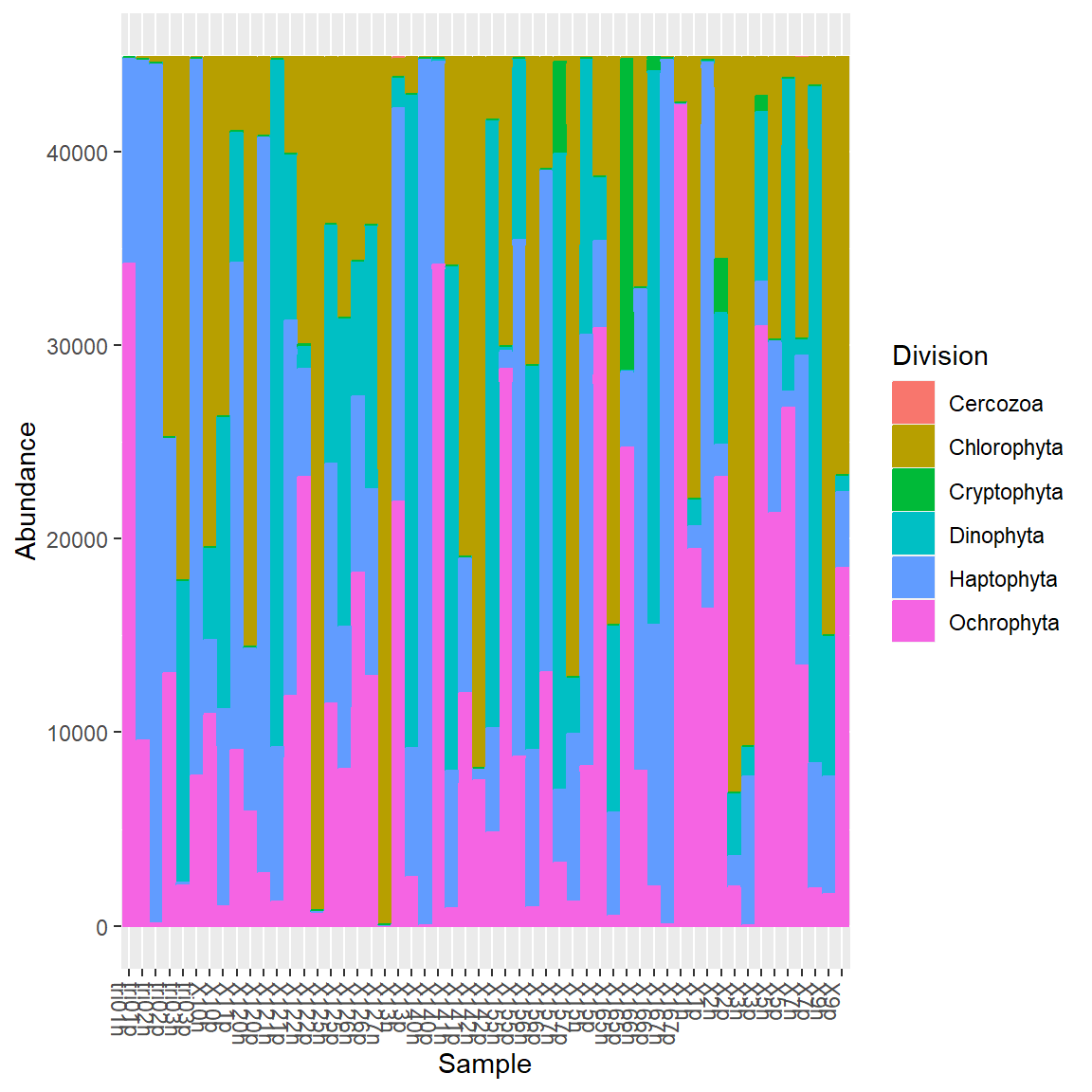
Remove OTUs boundaries.
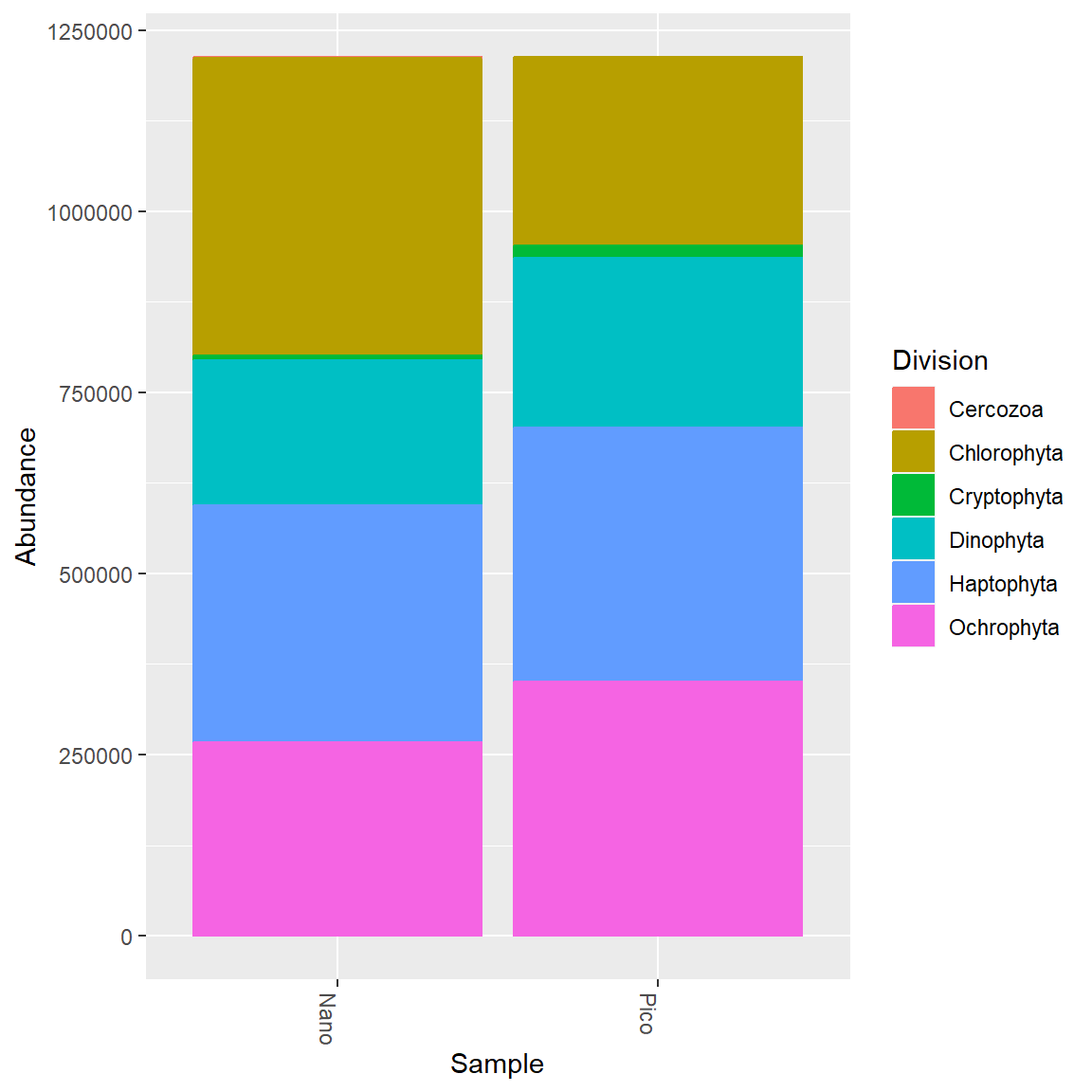
Group together Pico vs Nano samples.
More grouping
# Keep only Chlorophyta
# Color according to genus.
# Pico vs Nano/Surface vs Deep.
carbom_chloro_ps <- subset_taxa(carbom,
Division %in% c("Chlorophyta"))
# Transform from phylseq to dataframe
carbom_chloro_df <- psmelt(carbom_chloro_ps) %>%
# Group by fraction and level
group_by(fraction, level) %>%
# Compute relative % for each group
mutate(Abundance_pct = Abundance/sum(Abundance) * 100)
# Use ggplot directly
ggplot(carbom_chloro_df) +
geom_bar(aes(x= Division, y = Abundance_pct, fill=Genus),
stat="identity",
position="stack") +
facet_grid(rows = vars(level), cols=vars(fraction))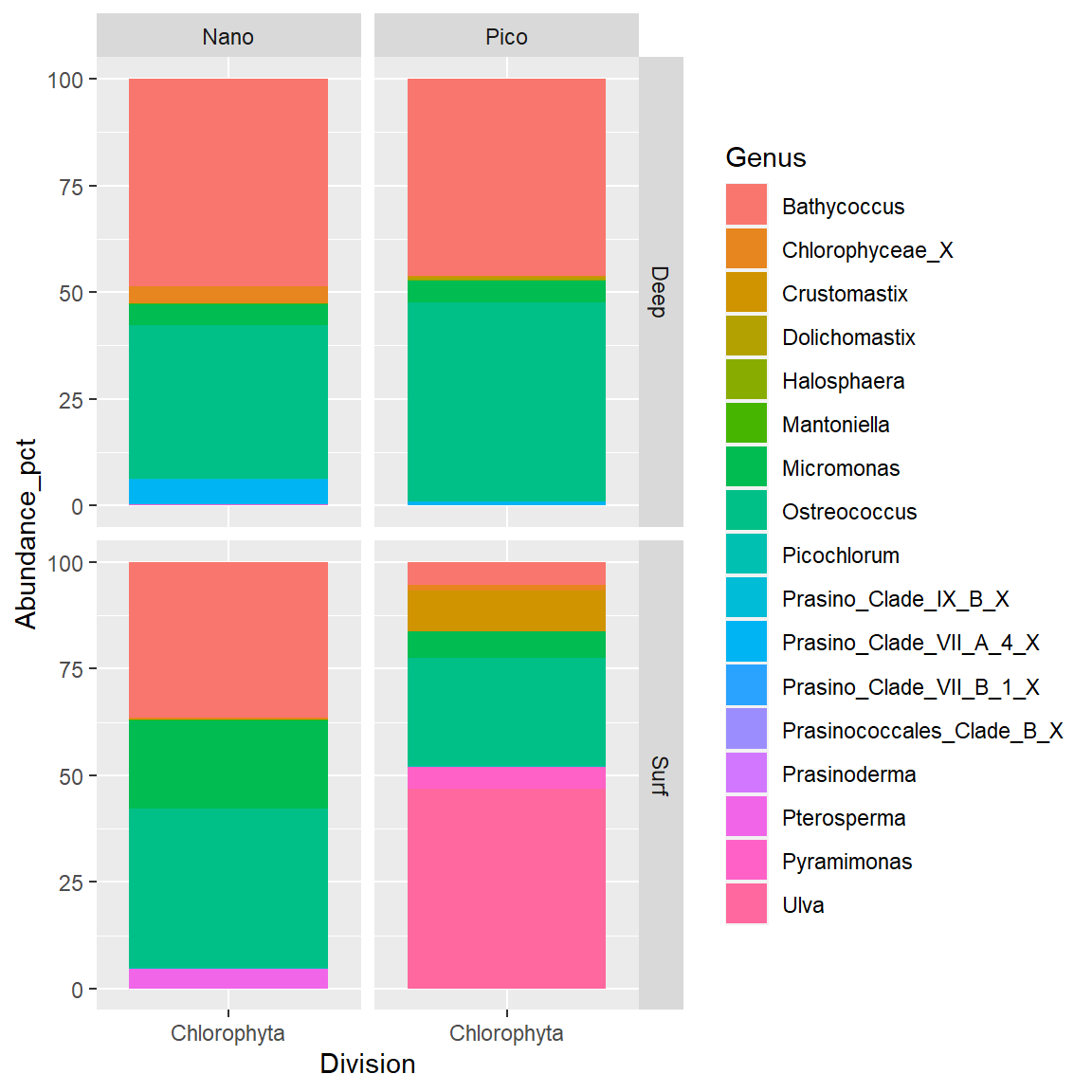
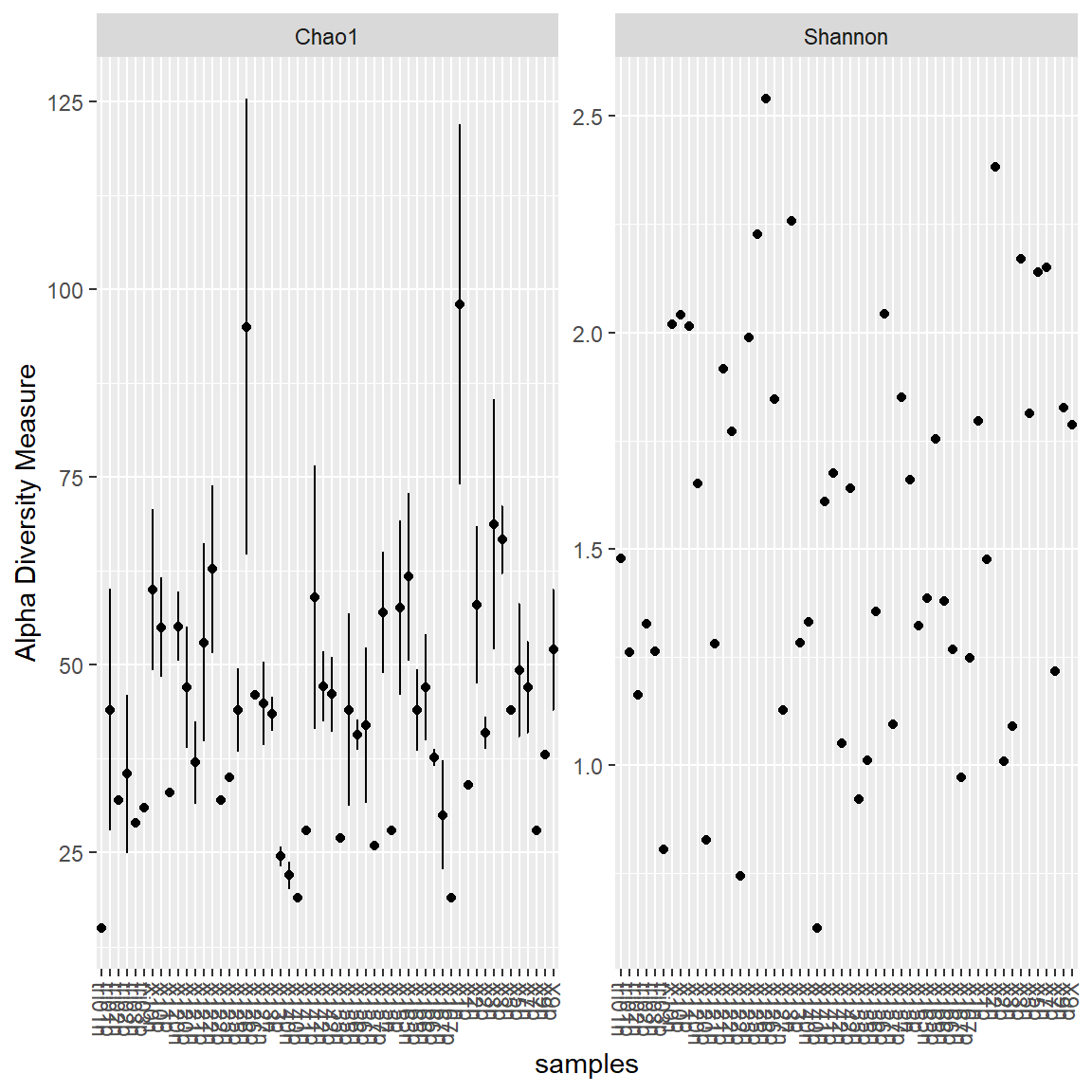
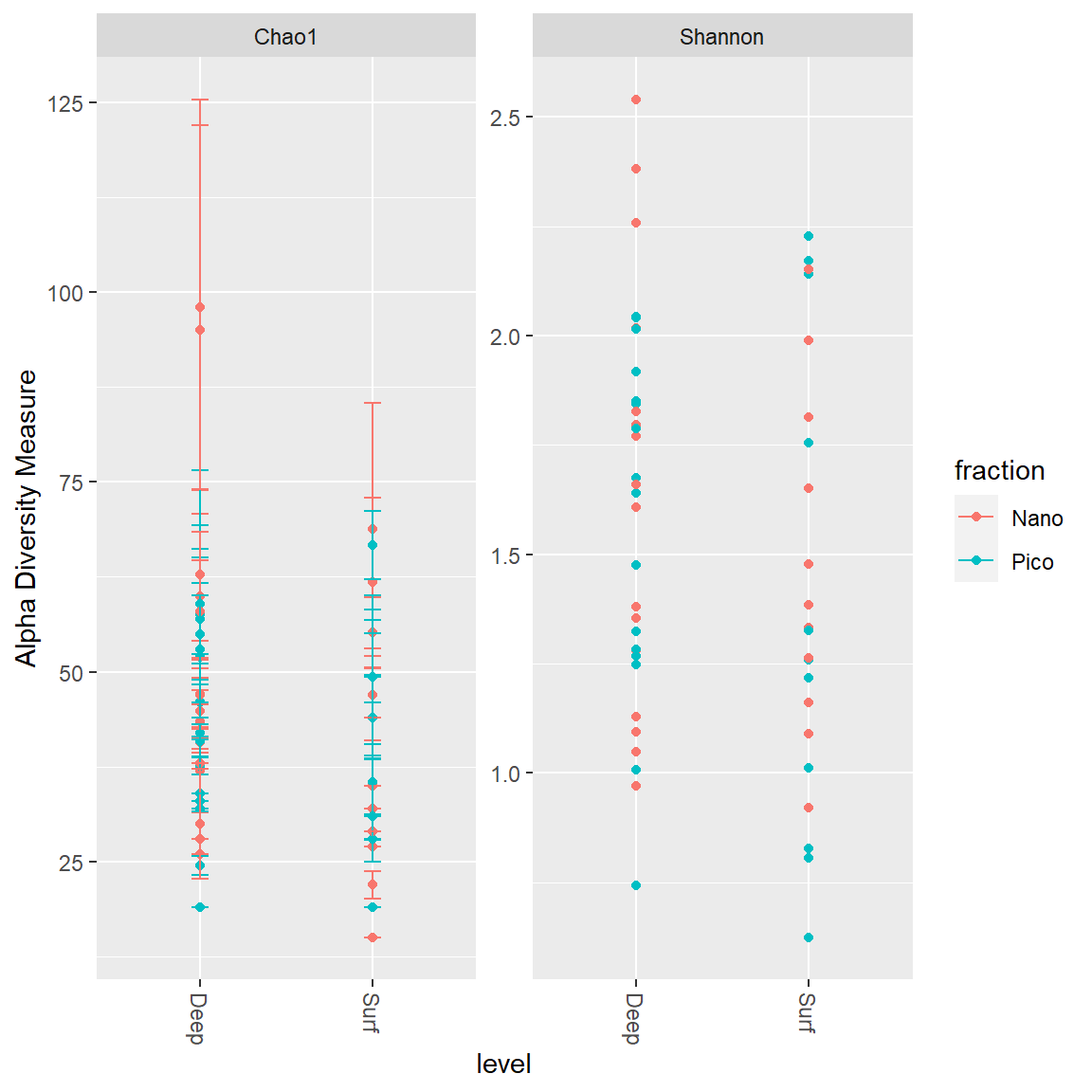
Alpha diversity
Diversity indexes
- Chao1: richness estimator
- Shannon: diversity estimator.
Other include: “Observed”, “ACE”, “Simpson”, “InvSimpson”, “Fisher”
Regroup together samples by level and fraction.
Beta diversity
NMDS
- Ordination: NMDS
- Distance: Bray
Square root transformation
Wisconsin double standardization
Run 0 stress 0.2317058
Run 1 stress 0.2554091
Run 2 stress 0.2541521
Run 3 stress 0.244086
Run 4 stress 0.2467867
Run 5 stress 0.2515843
Run 6 stress 0.2458829
Run 7 stress 0.2588258
Run 8 stress 0.2399245
Run 9 stress 0.2526569
Run 10 stress 0.2460093
Run 11 stress 0.2508037
Run 12 stress 0.2618731
Run 13 stress 0.2412877
Run 14 stress 0.2482059
Run 15 stress 0.2294626
... New best solution
... Procrustes: rmse 0.09894239 max resid 0.3902299
Run 16 stress 0.2541231
Run 17 stress 0.2486373
Run 18 stress 0.2478466
Run 19 stress 0.2508294
Run 20 stress 0.2316124
*** Best solution was not repeated -- monoMDS stopping criteria:
18: stress ratio > sratmax
2: scale factor of the gradient < sfgrminPlot OTUs
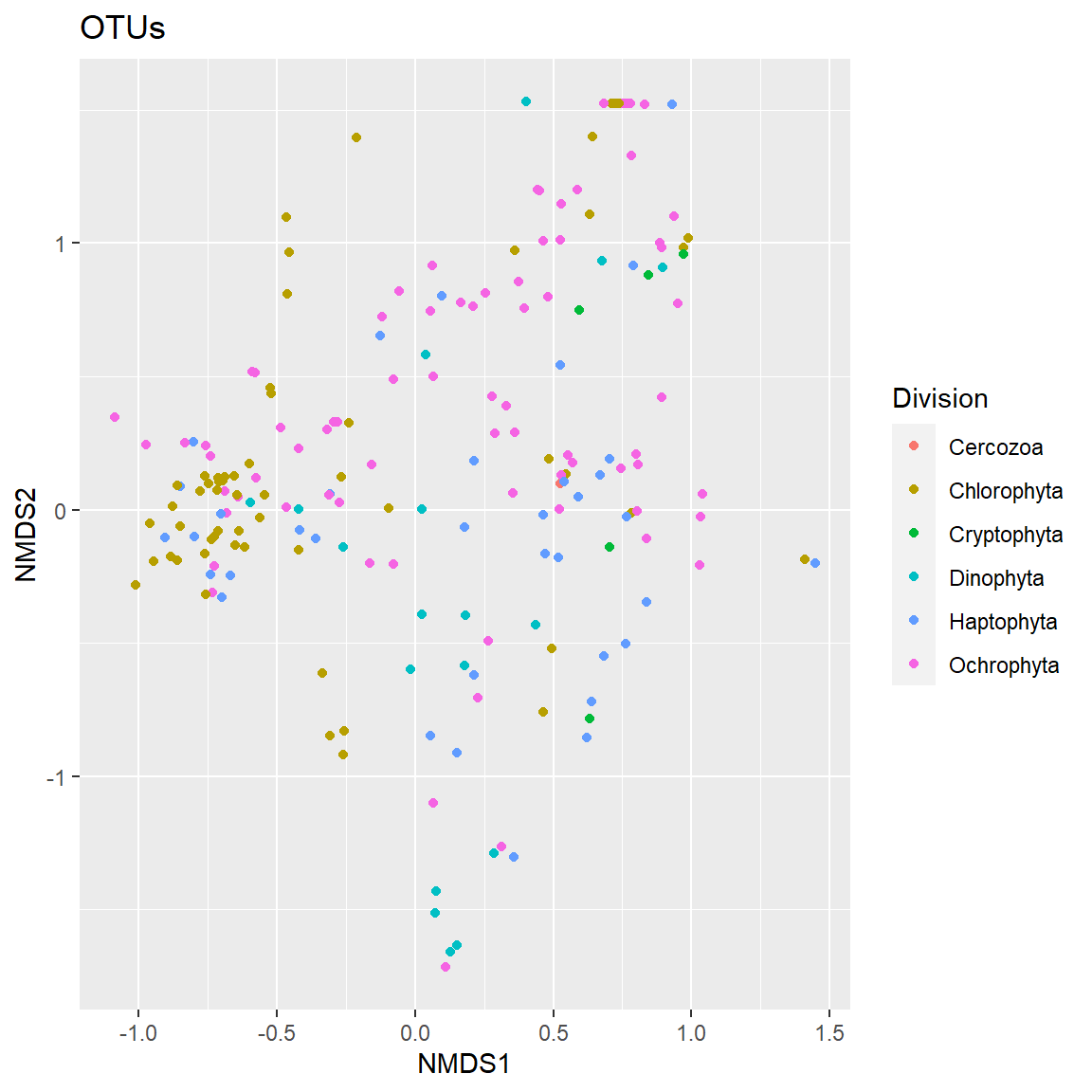
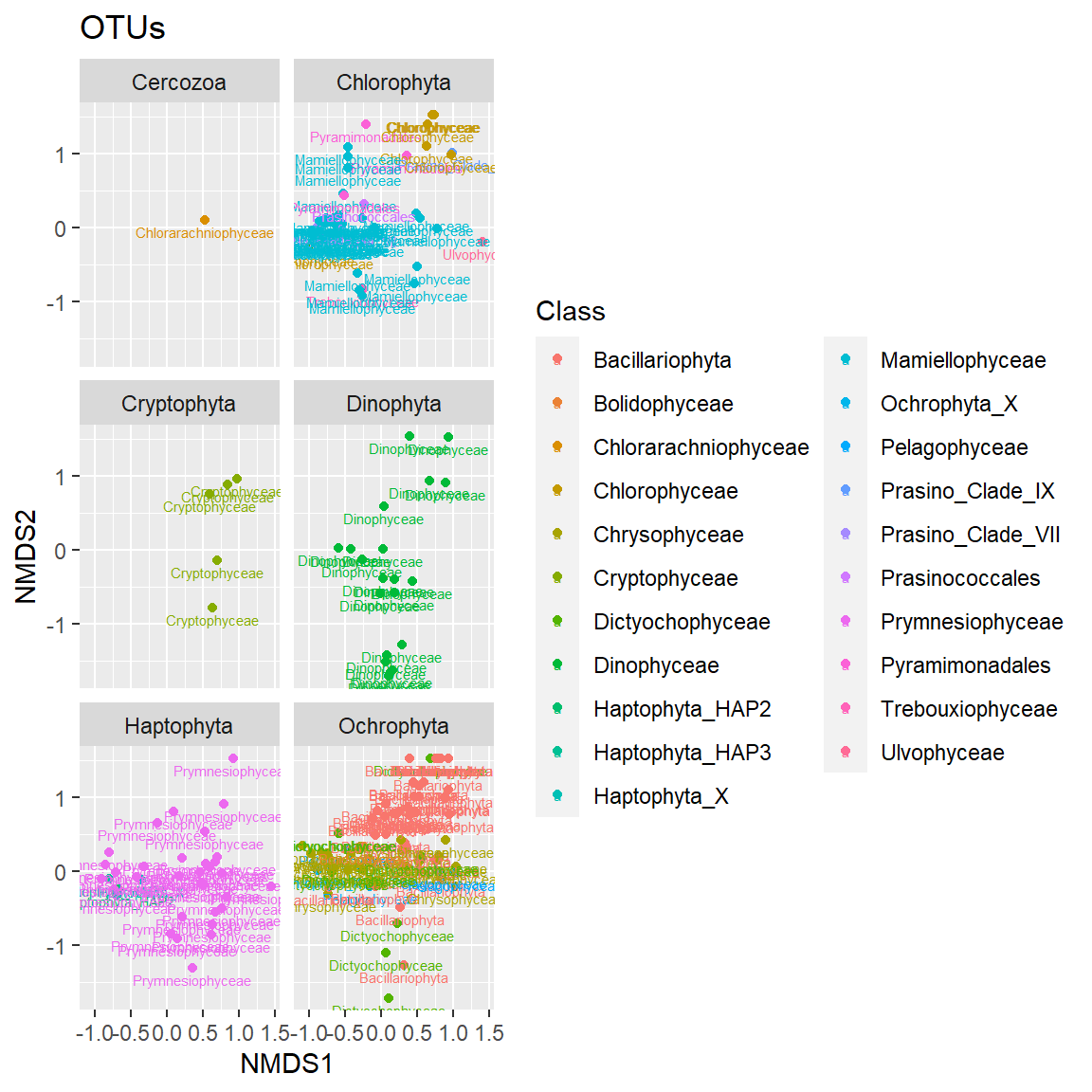
Plot OTUs
A bit confusing, so make it more easy to visualize by breaking according to taxonomic division.
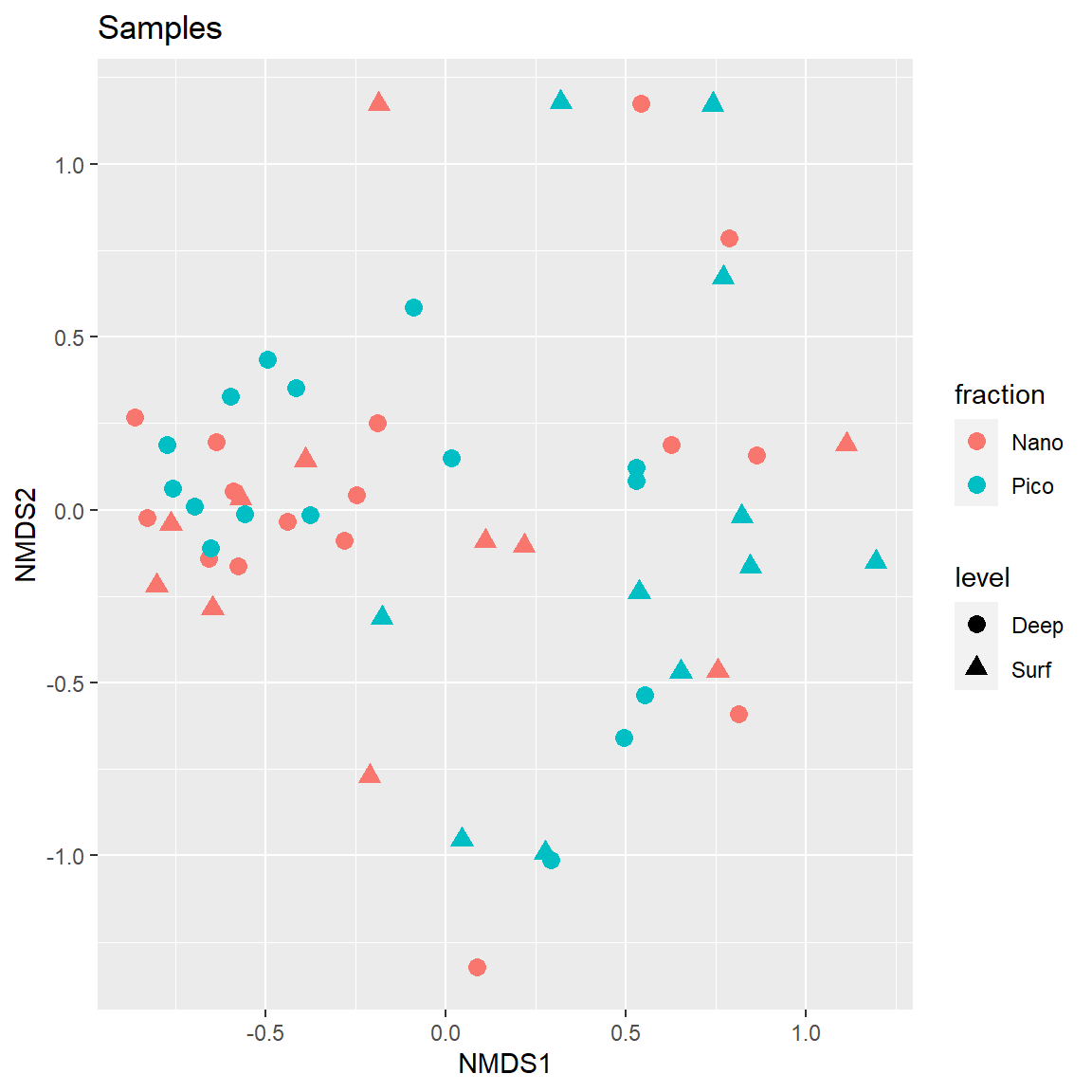
Plot Samples
Enlarge the points to make it more easy to read.
Plot both OTUs and Samples
Two different panels
Methods
CCA - Performs correspondence analysis, or optionally, constrained correspondence analysis (a.k.a. canonical correspondence analysis), via cca
RDA - Performs redundancy analysis, or optionally principal components analysis, via rda
NMDS - Performs Non-metric MultiDimenstional Scaling of a sample-wise ecological distance matrix onto a user-specified number of axes, k. By default, k=2, but this can be modified as a supplementary argument.
MDS/PCoA - Performs principal coordinate analysis (also called principle coordinate decomposition, multidimensional scaling (MDS), or classical scaling) of a distance matrix (Gower 1966), including two correction methods for negative eigenvalues. See pcoa for further details.
Distances
- manhattan
- euclidean
- canberra
- bray
- kulczynski
- jaccard
- gower
- altGower
- morisita
- horn
- mountford
- raup
- binomial
- chao
- cao
- unifrac
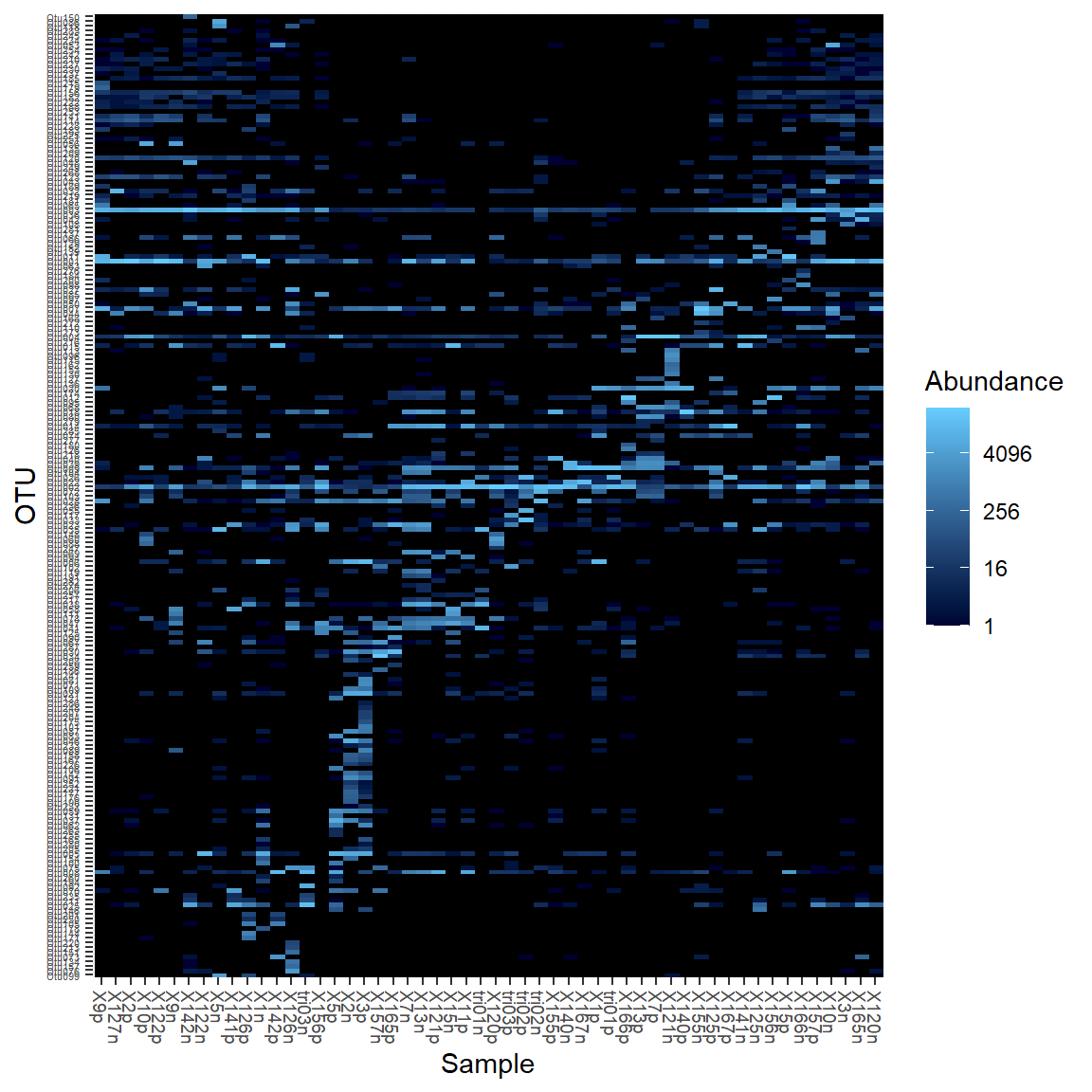
Heatmaps
Basic heatmap using the default parameters.
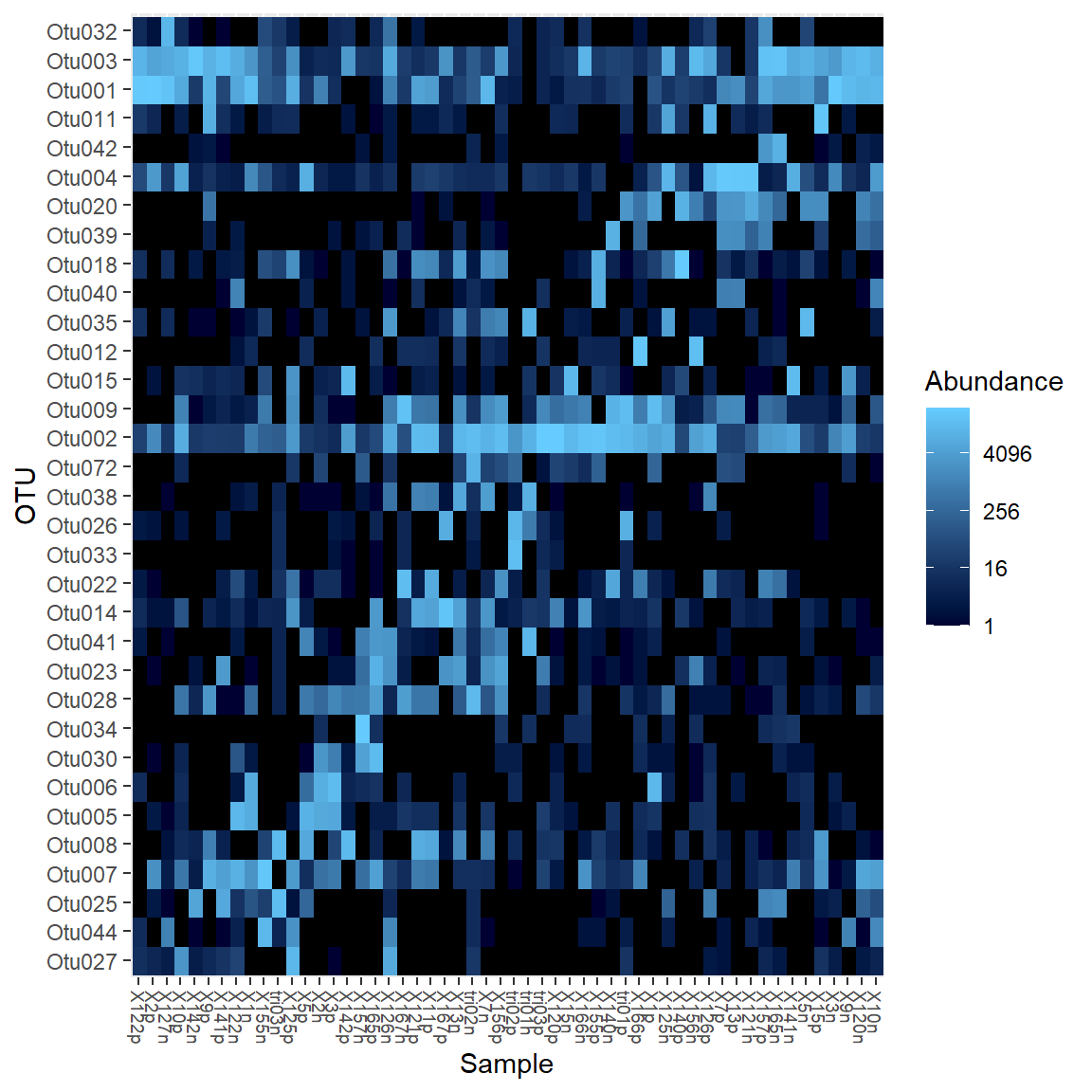
Consider only main OTUS
It is very very cluttered. It is better to only consider the most abundant OTUs for heatmaps. For example one can only take OTUs that represent at least 20% of reads in at least one sample. Remember we normalized all the sampples to median number of reads (total). We are left with only 33 OTUS which makes the reading much more easy.
phyloseq-class experiment-level object
otu_table() OTU Table: [ 33 taxa and 54 samples ]
sample_data() Sample Data: [ 54 samples by 27 sample variables ]
tax_table() Taxonomy Table: [ 33 taxa by 7 taxonomic ranks ]OTU Table: [8 taxa and 5 samples]
taxa are rows
X10n X10p X11p X120n X120p
Otu001 13339 7346 3804 12662 3
Otu002 18 8329 14958 30 36206
Otu003 9692 10488 20 16537 11
Otu004 3584 4943 33 7 9
Otu005 0 6 11 0 5
Otu006 0 9 0 0 5
Otu007 4473 605 587 5894 3
Otu008 1 9 6707 2 17Consider only main OTUS
Change distances
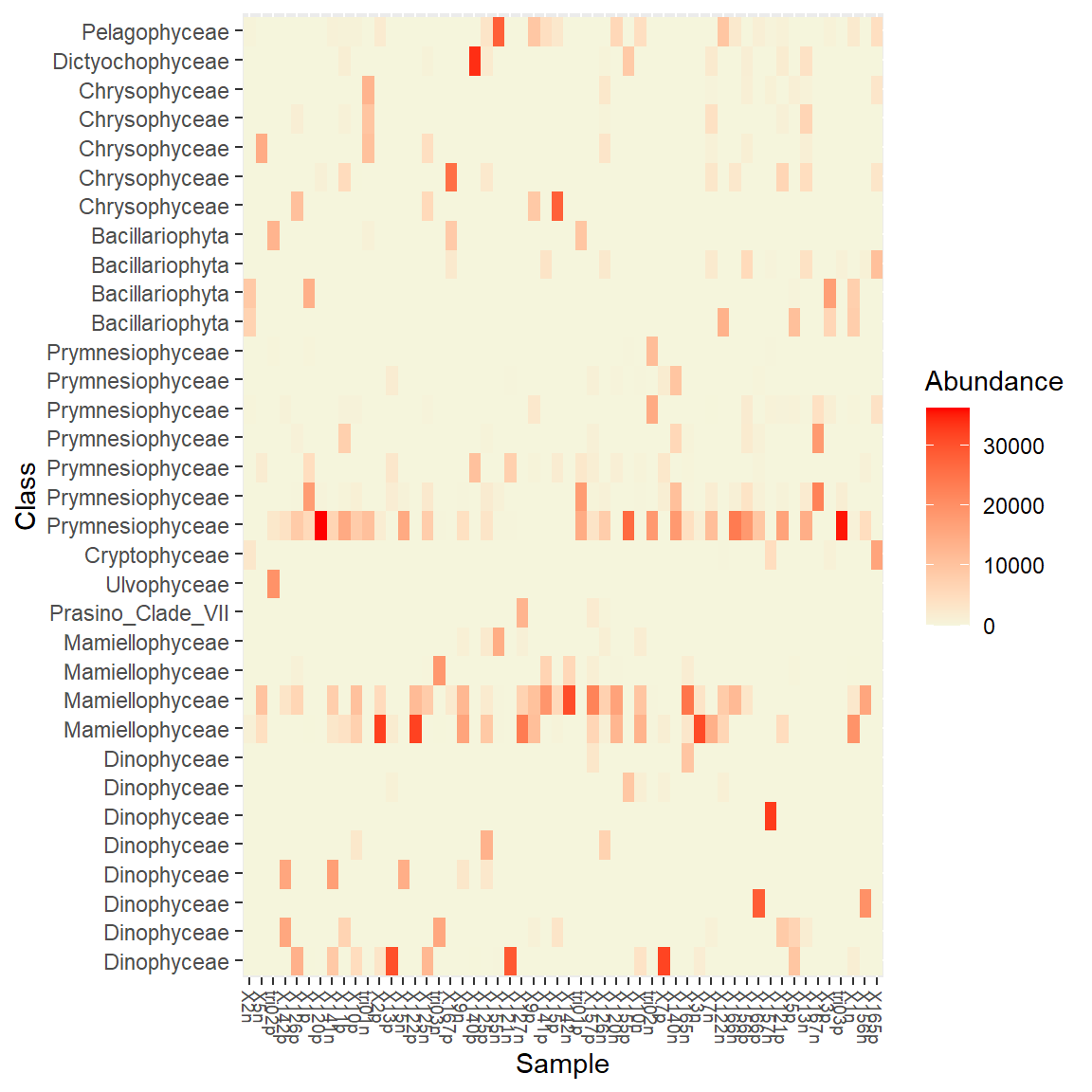
It is possible to use different distances and different multivaraite methods. For example Jaccard distance and MDS and label OTUs with Class, order by Class. We can also change the Palette (the default palette is a bit ugly…).
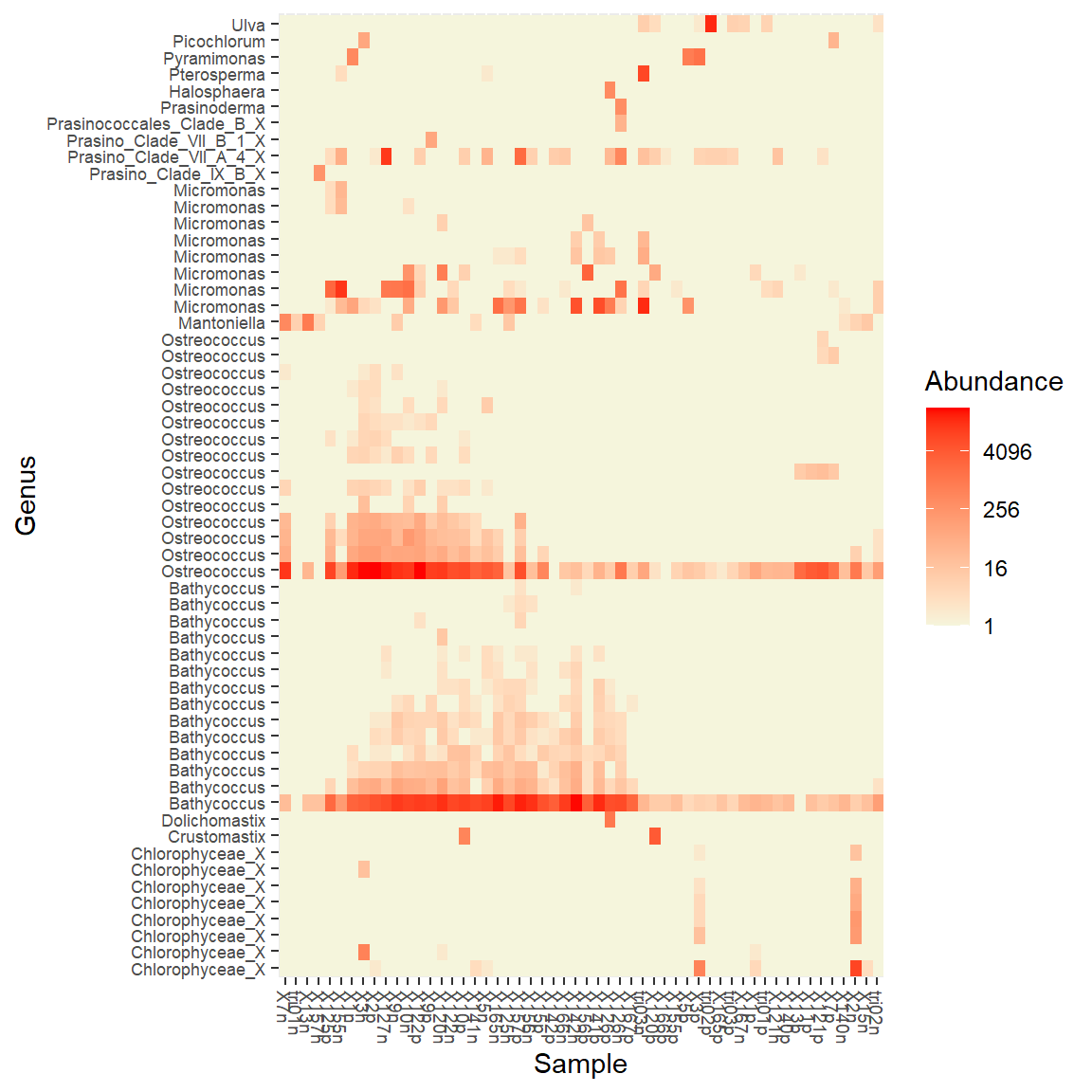
Heatmap for a specific taxonomy group.
For example we can taget the Chlorophyta and then label the OTUs using the Genus.
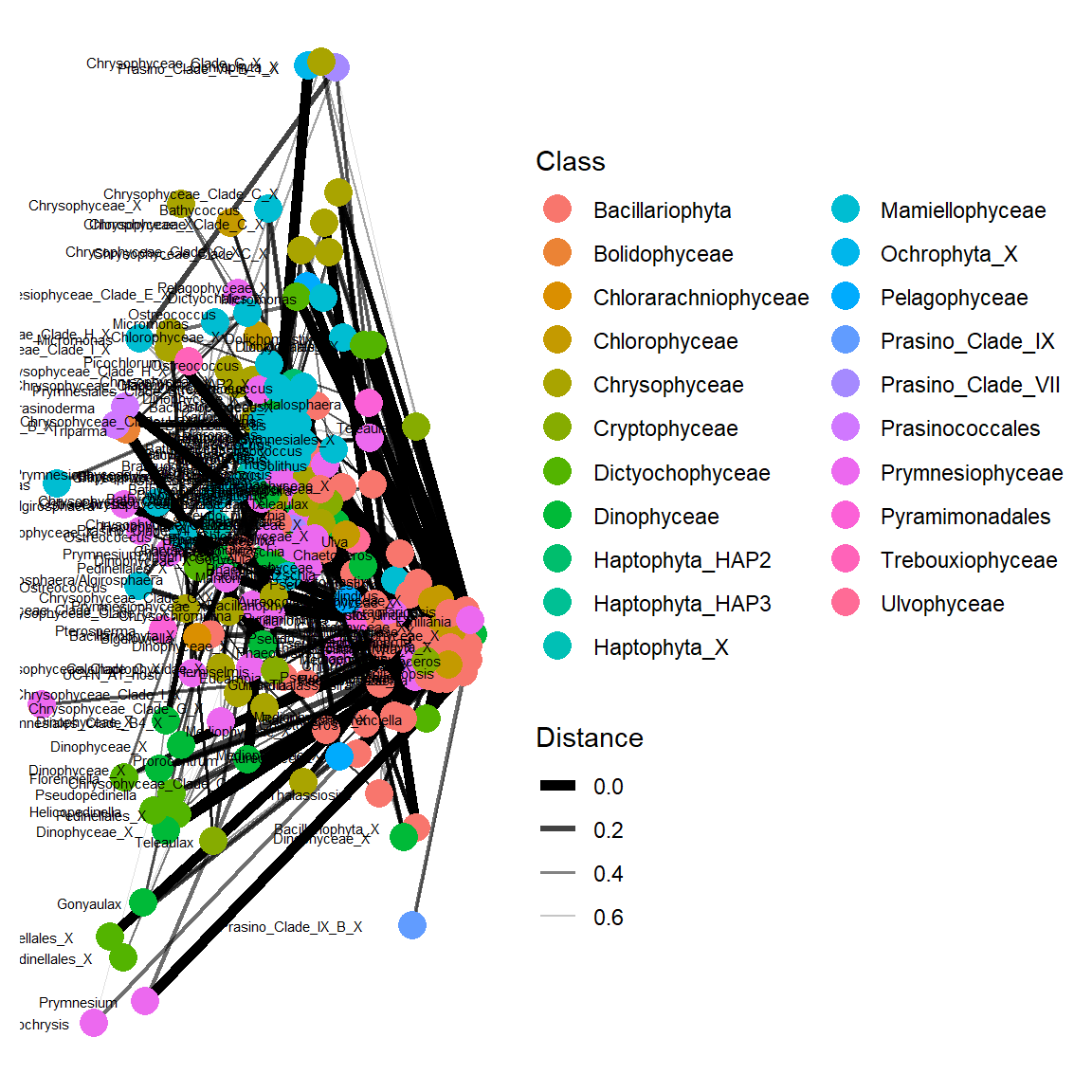
Networks
Simple network analysis
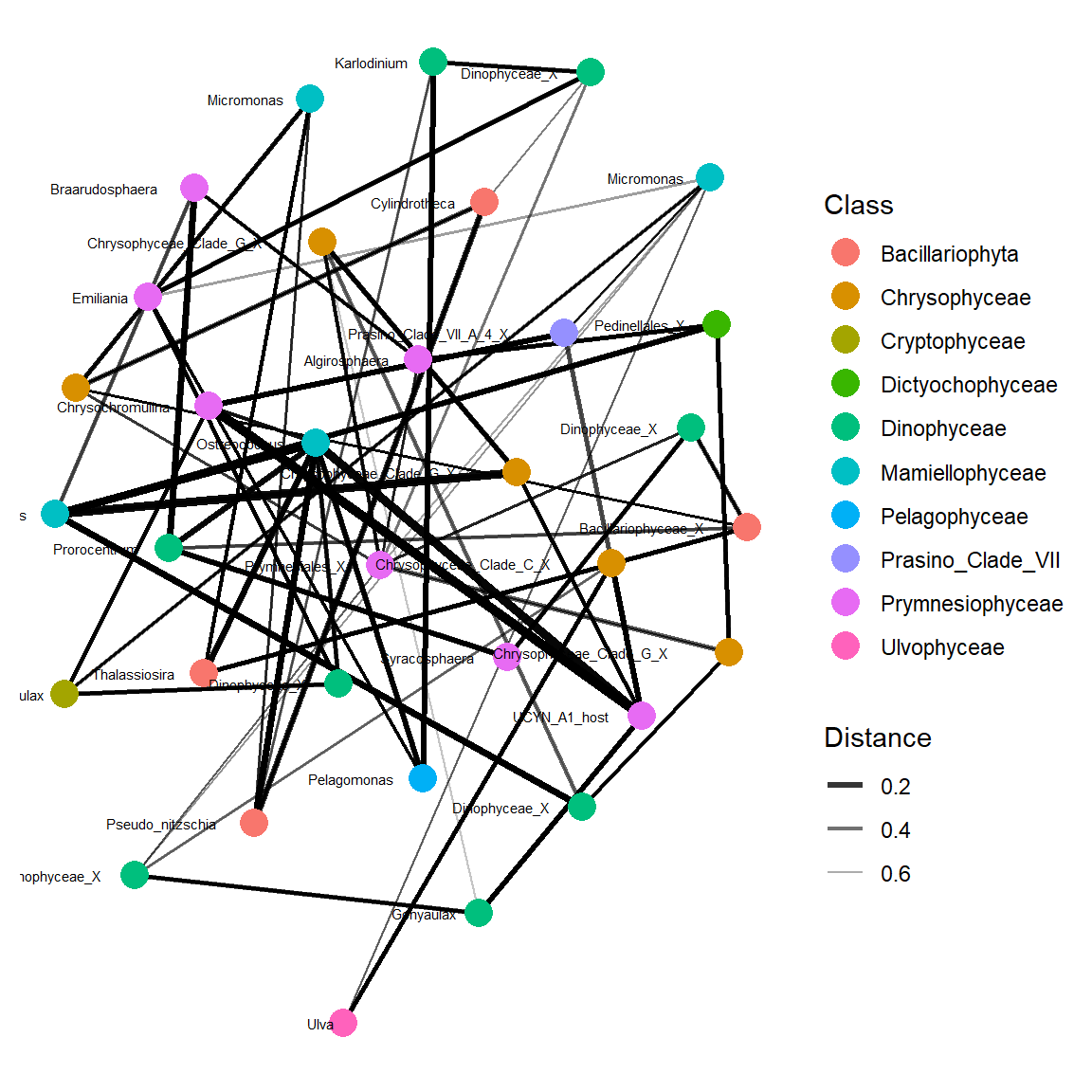
Simplify
Let us make it more simple by using only major OTUs
What did we learn ?
- Create file
- Bar graphs
- Alpha diversity
- Beta diversity
R - Phyloseq