R course
Daniel Vaulot
2023-01-19



Data wrangling
R - Session 02
- Data frames
- Concept of tidy data
- Reading data
- Manipulating data
- Columns
- Rows
Data frames
R objects
List
Matrix
Factors
Data frames
Data frames
What is it ?
- Table mixing different types of columns (an Excel table…)
- However within a column all values are similar, e.g. numeric, logical, character
label id value flag
1 a 1 0.03963652 TRUE
2 b 2 -1.03193245 FALSE
3 c 3 -0.46903884 TRUE
4 d 4 -1.36385849 FALSE
5 e 5 0.71374085 TRUE
6 f 6 0.48473275 FALSE* We will NOT use factors: stringsAsFactors = FALSE (default in R > 4.0)
Useful functions
[1] 6 4
[1] 6
[1] 4'data.frame': 6 obs. of 4 variables:
$ label: chr "a" "b" "c" "d" ...
$ id : int 1 2 3 4 5 6
$ value: num 0.0396 -1.0319 -0.469 -1.3639 0.7137 ...
$ flag : logi TRUE FALSE TRUE FALSE TRUE FALSE
[1] "label" "id" "value" "flag" Access specific value
- Use the
df[i,j]notation, first index corresponds to row, second index to column
[1] 0.7137409Access specific column
- Use the
df[i,j]notation ::: {.cell output-location=‘fragment’}
[1] 0.03963652 -1.03193245 -0.46903884 -1.36385849 0.71374085 0.48473275
[1] 0.03963652 -1.03193245 -0.46903884 -1.36385849 0.71374085 0.48473275::: The result is a vector
Access row
- Use the
df[i,j]notation ::: {.cell output-location=‘fragment’}
label id value flag
1 a 1 0.03963652 TRUE:::
The result is a data frame
Access specific rows
- e.g. Rows for which the value of id <= 3
label id value flag
1 a 1 0.03963652 TRUE
2 b 2 -1.03193245 FALSE
3 c 3 -0.46903884 TRUESelect lines for which the label is c
This syntax is complicated - tidyverse packages make it much more easy to manipulate and remember
Tidy data
Installation and Resources
Packages
- readxl : Reading Excel files
- readr : Reading and writing Text files
- dplyr : Filter and reformat data frames
- tidyr : Make data “tidy”
- stringr : Manipulating strings
- lubridate : Manipulate date
Data and script
- unzip
data.zip - Open in R
scripts/script_wrangling.R
Resources
R for data science: (Chapter 5)
Cheat sheets
Basic concepts
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.

Load necessary libraries
Read and Write data
Oceanographic data
CARBOM cruise off Brazil
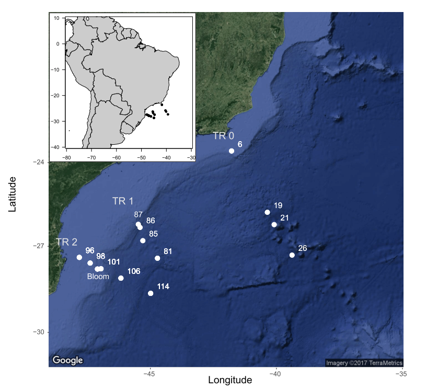
- Stations
- Depth
- Coordinates
- Temperature, Salinity
- Nitrates, Phosphates
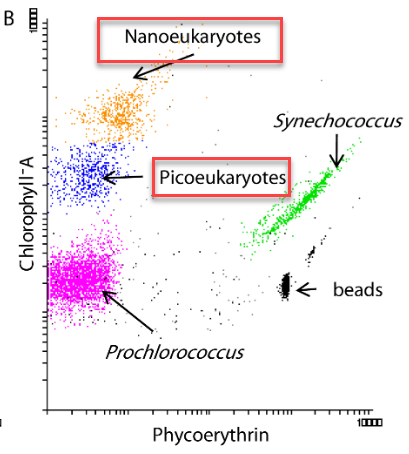
Microbial populations
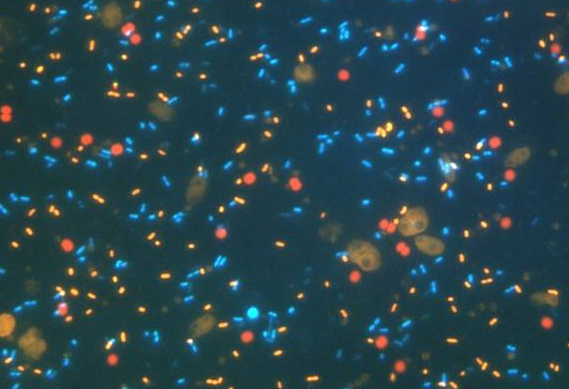
- Flow cytometry :
- pico-eukaryotes
- nano-eukaryotes
Read data
Text file - TAB delimited

Reading a text file
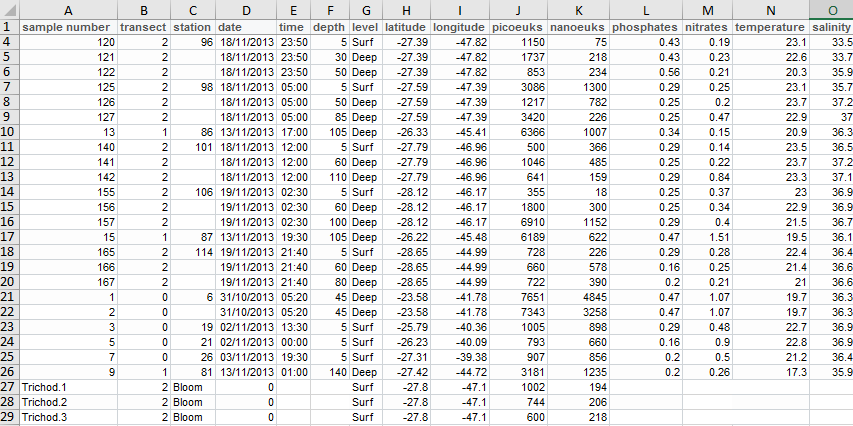
| sample number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 13/11/2013 | 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 |
| 11 | 1 | 85 | 13/11/2013 | 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 |
| 120 | 2 | 96 | 18/11/2013 | 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 |
| 121 | 2 | 18/11/2013 | 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 | |
| 122 | 2 | 18/11/2013 | 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 | |
| 125 | 2 | 98 | 18/11/2013 | 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 |
| 126 | 2 | 18/11/2013 | 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 | |
| 127 | 2 | 18/11/2013 | 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 | |
| 13 | 1 | 86 | 13/11/2013 | 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 |
| 140 | 2 | 101 | 18/11/2013 | 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 |
readr::read_tsv() : read tab delimited files
readr::read_csv() : read comma delimited files
readr::write_tsv() : write tab delimited files
Excel sheet
Read the data - read_excel
| sample number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 |
| 121 | 2 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 | |
| 122 | 2 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 | |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 |
| 126 | 2 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 | |
| 127 | 2 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 | |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 |
- Can also select a range : e.g. A1:Q26
- Can skip lines
Bad data input under Excel
| sample number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 |
| 121 | 2 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 | |
| 122 | 2 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 | |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 |
| 126 | 2 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 | |
| 127 | 2 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 | |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 |
- There are missing values in the column station because only recorded when changed
Filling missing values - fill
| sample number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 |
| 121 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 |
| 122 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 |
| 126 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 |
| 127 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 |
- All missing values have been filled in.
Write data
Text file
- readr::write_tsv() : write tab delimited files
Excel file
openxlsx::write.xlsx : write tab delimited files
Many options: specific sheet, formatting etc…
Write data
Library rio
- Many output formats
- import() / export()
dplyr - Manipulate tables

@allison_horst
Manipulate columns
List and Summarize columns
List columns
[1] "sample number" "transect" "station" "date"
[5] "time" "depth" "level" "latitude"
[9] "longitude" "picoeuks" "nanoeuks" "phosphates"
[13] "nitrates" "temperature" "salinity" Select specific columns - select
| transect | sample number | station | depth | latitude | longitude | picoeuks | nanoeuks |
|---|---|---|---|---|---|---|---|
| 1 | 10 | 81 | 140 | -27.42 | -44.72 | 3278 | 1232 |
| 1 | 11 | 85 | 110 | -26.80 | -45.30 | 16312 | 1615 |
| 2 | 120 | 96 | 5 | -27.39 | -47.82 | 1150 | 75 |
| 2 | 121 | 96 | 30 | -27.39 | -47.82 | 1737 | 218 |
| 2 | 122 | 96 | 50 | -27.39 | -47.82 | 853 | 234 |
| 2 | 125 | 98 | 5 | -27.59 | -47.39 | 3086 | 1300 |
| 2 | 126 | 98 | 50 | -27.59 | -47.39 | 1217 | 782 |
| 2 | 127 | 98 | 85 | -27.59 | -47.39 | 3420 | 226 |
| 1 | 13 | 86 | 105 | -26.33 | -45.41 | 6366 | 1007 |
| 2 | 140 | 101 | 5 | -27.79 | -46.96 | 500 | 366 |
* Column names are not “quoted” (in base R you need to “quote” the column names)
* Better not to put space in column header because then must enclose column name with ` (back-quote)
Select a range of columns - select
| transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 |
| 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 |
| 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 |
| 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 |
| 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 |
| 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 |
| 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 |
| 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 |
| 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 |
| 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 |
Unselect columns - select
| sample number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | temperature | salinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 17.3 | 35.9 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 21.3 | 36.5 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 23.1 | 33.5 |
| 121 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 22.6 | 33.7 |
| 122 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 20.3 | 35.9 |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 23.1 | 35.7 |
| 126 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 23.7 | 37.2 |
| 127 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 22.9 | 37.0 |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 20.9 | 36.3 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 23.5 | 36.5 |
Using the pipe operator - %>%
| transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 |
| 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 |
| 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 |
| 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 |
| 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 |
| 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 |
| 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 |
| 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 |
| 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 |
| 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 |
Renaming variables - rename
| sample_number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 |
| 121 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 |
| 122 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 |
| 126 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 |
| 127 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 |
Creating new variables - mutate
| sample_number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity | pico_pct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 | 72.68293 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 | 90.99124 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 | 93.87755 |
| 121 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 | 88.84910 |
| 122 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 | 78.47286 |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 | 70.36024 |
| 126 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 | 60.88044 |
| 127 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 | 93.80143 |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 | 86.34206 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 | 57.73672 |
- You can also use transmute() but then it will drop all the other columns.
- It is much much better to compute new variables in R than in Excel, because you can easily track and correct errors.
Using the pipe operator you can chain operations
| sample_number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | pico_pct |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 72.68293 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 90.99124 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 93.87755 |
| 121 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 88.84910 |
| 122 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 78.47286 |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 70.36024 |
| 126 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 60.88044 |
| 127 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 93.80143 |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 86.34206 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 57.73672 |
Creating labels with mutate and stringr functions
| sample_number | transect | station | date | time | sample_label |
|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1899-12-31 01:00:00 | TR_1_St_81 |
| 11 | 1 | 85 | 2013-11-13 | 1899-12-31 13:30:00 | TR_1_St_85 |
| 120 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | TR_2_St_96 |
| 121 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | TR_2_St_96 |
| 122 | 2 | 96 | 2013-11-18 | 1899-12-31 23:50:00 | TR_2_St_96 |
| 125 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | TR_2_St_98 |
| 126 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | TR_2_St_98 |
| 127 | 2 | 98 | 2013-11-18 | 1899-12-31 05:00:00 | TR_2_St_98 |
| 13 | 1 | 86 | 2013-11-13 | 1899-12-31 17:00:00 | TR_1_St_86 |
| 140 | 2 | 101 | 2013-11-18 | 1899-12-31 12:00:00 | TR_2_St_101 |
Changing type of some columns - mutate
- Use the
lubridatepackage to manipulate dates
| sample_number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity | pico_pct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 1 | 81 | 2013-11-13 | 1:0 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 | 72.68293 |
| 11 | 1 | 85 | 2013-11-13 | 13:30 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 | 90.99124 |
| 120 | 2 | 96 | 2013-11-18 | 23:50 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 | 93.87755 |
| 121 | 2 | 96 | 2013-11-18 | 23:50 | 30 | Deep | -27.39 | -47.82 | 1737 | 218 | 0.43 | 0.23 | 22.6 | 33.7 | 88.84910 |
| 122 | 2 | 96 | 2013-11-18 | 23:50 | 50 | Deep | -27.39 | -47.82 | 853 | 234 | 0.56 | 0.21 | 20.3 | 35.9 | 78.47286 |
| 125 | 2 | 98 | 2013-11-18 | 5:0 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 | 70.36024 |
| 126 | 2 | 98 | 2013-11-18 | 5:0 | 50 | Deep | -27.59 | -47.39 | 1217 | 782 | 0.25 | 0.20 | 23.7 | 37.2 | 60.88044 |
| 127 | 2 | 98 | 2013-11-18 | 5:0 | 85 | Deep | -27.59 | -47.39 | 3420 | 226 | 0.25 | 0.47 | 22.9 | 37.0 | 93.80143 |
| 13 | 1 | 86 | 2013-11-13 | 17:0 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 | 86.34206 |
| 140 | 2 | 101 | 2013-11-18 | 12:0 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 | 57.73672 |
Manipulating rows
Order rows - arrange
| sample_number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity | pico_pct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 0 | 19 | 2013-11-02 | 13:30 | 5 | Surf | -25.79 | -40.36 | 1005 | 898 | 0.29 | 0.48 | 22.7 | 36.9 | 52.81135 |
| 5 | 0 | 21 | 2013-11-02 | 0:0 | 5 | Surf | -26.23 | -40.09 | 793 | 660 | 0.16 | 0.90 | 22.8 | 36.9 | 54.57674 |
| 7 | 0 | 26 | 2013-11-03 | 19:30 | 5 | Surf | -27.31 | -39.38 | 907 | 856 | 0.20 | 0.50 | 21.2 | 36.4 | 51.44640 |
| 1 | 0 | 6 | 2013-10-31 | 5:20 | 45 | Deep | -23.58 | -41.78 | 7651 | 4845 | 0.47 | 1.07 | 19.7 | 36.3 | 61.22759 |
| 2 | 0 | 6 | 2013-10-31 | 5:20 | 45 | Deep | -23.58 | -41.78 | 7343 | 3258 | 0.47 | 1.07 | 19.7 | 36.3 | 69.26705 |
| 10 | 1 | 81 | 2013-11-13 | 1:0 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 | 72.68293 |
| 9 | 1 | 81 | 2013-11-13 | 1:0 | 140 | Deep | -27.42 | -44.72 | 3181 | 1235 | 0.20 | 0.26 | 17.3 | 35.9 | 72.03351 |
| 11 | 1 | 85 | 2013-11-13 | 13:30 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 | 90.99124 |
| 13 | 1 | 86 | 2013-11-13 | 17:0 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 | 86.34206 |
| 15 | 1 | 87 | 2013-11-13 | 19:30 | 105 | Deep | -26.22 | -45.48 | 6189 | 622 | 0.47 | 1.51 | 19.5 | 36.1 | 90.86771 |
- Station 6 is not ordered numerically. It is because station is a character column.
Order rows - transform to numeric
| sample_number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity | pico_pct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 6 | 2013-10-31 | 5:20 | 45 | Deep | -23.58 | -41.78 | 7651 | 4845 | 0.47 | 1.07 | 19.7 | 36.3 | 61.22759 |
| 2 | 0 | 6 | 2013-10-31 | 5:20 | 45 | Deep | -23.58 | -41.78 | 7343 | 3258 | 0.47 | 1.07 | 19.7 | 36.3 | 69.26705 |
| 3 | 0 | 19 | 2013-11-02 | 13:30 | 5 | Surf | -25.79 | -40.36 | 1005 | 898 | 0.29 | 0.48 | 22.7 | 36.9 | 52.81135 |
| 5 | 0 | 21 | 2013-11-02 | 0:0 | 5 | Surf | -26.23 | -40.09 | 793 | 660 | 0.16 | 0.90 | 22.8 | 36.9 | 54.57674 |
| 7 | 0 | 26 | 2013-11-03 | 19:30 | 5 | Surf | -27.31 | -39.38 | 907 | 856 | 0.20 | 0.50 | 21.2 | 36.4 | 51.44640 |
| 10 | 1 | 81 | 2013-11-13 | 1:0 | 140 | Deep | -27.42 | -44.72 | 3278 | 1232 | 0.20 | 0.26 | 17.3 | 35.9 | 72.68293 |
| 9 | 1 | 81 | 2013-11-13 | 1:0 | 140 | Deep | -27.42 | -44.72 | 3181 | 1235 | 0.20 | 0.26 | 17.3 | 35.9 | 72.03351 |
| 11 | 1 | 85 | 2013-11-13 | 13:30 | 110 | Deep | -26.80 | -45.30 | 16312 | 1615 | 0.29 | 0.22 | 21.3 | 36.5 | 90.99124 |
| 13 | 1 | 86 | 2013-11-13 | 17:0 | 105 | Deep | -26.33 | -45.41 | 6366 | 1007 | 0.34 | 0.15 | 20.9 | 36.3 | 86.34206 |
| 15 | 1 | 87 | 2013-11-13 | 19:30 | 105 | Deep | -26.22 | -45.48 | 6189 | 622 | 0.47 | 1.51 | 19.5 | 36.1 | 90.86771 |
- One station named “Bloom” could not be converted to numerical (-> NA)
Summarize rows - count
- Compute number of stations per transect
| transect | n |
|---|---|
| 0 | 5 |
| 1 | 5 |
| 2 | 18 |
Summarize rows - group_by / summarize
- Group by transect and station
- Compute mean of the percent picoplankton
| transect | station | n_samples | mean_pico_percent |
|---|---|---|---|
| 0 | 6 | 2 | 65.24732 |
| 0 | 19 | 1 | 52.81135 |
| 0 | 21 | 1 | 54.57674 |
| 0 | 26 | 1 | 51.44640 |
| 1 | 81 | 2 | 72.35822 |
| 1 | 85 | 1 | 90.99124 |
| 1 | 86 | 1 | 86.34206 |
| 1 | 87 | 1 | 90.86771 |
| 2 | 96 | 3 | 87.06651 |
| 2 | 98 | 3 | 75.01403 |
Filtering rows - filter
- Get only the surface samples
| sample_number | transect | station | date | time | depth | level | latitude | longitude | picoeuks | nanoeuks | phosphates | nitrates | temperature | salinity | pico_pct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 0 | 19 | 2013-11-02 | 13:30 | 5 | Surf | -25.79 | -40.36 | 1005 | 898 | 0.29 | 0.48 | 22.7 | 36.9 | 52.81135 |
| 5 | 0 | 21 | 2013-11-02 | 0:0 | 5 | Surf | -26.23 | -40.09 | 793 | 660 | 0.16 | 0.90 | 22.8 | 36.9 | 54.57674 |
| 7 | 0 | 26 | 2013-11-03 | 19:30 | 5 | Surf | -27.31 | -39.38 | 907 | 856 | 0.20 | 0.50 | 21.2 | 36.4 | 51.44640 |
| 120 | 2 | 96 | 2013-11-18 | 23:50 | 5 | Surf | -27.39 | -47.82 | 1150 | 75 | 0.43 | 0.19 | 23.1 | 33.5 | 93.87755 |
| 125 | 2 | 98 | 2013-11-18 | 5:0 | 5 | Surf | -27.59 | -47.39 | 3086 | 1300 | 0.29 | 0.25 | 23.1 | 35.7 | 70.36024 |
| 140 | 2 | 101 | 2013-11-18 | 12:0 | 5 | Surf | -27.79 | -46.96 | 500 | 366 | 0.29 | 0.14 | 23.5 | 36.5 | 57.73672 |
| 155 | 2 | 106 | 2013-11-19 | 2:30 | 5 | Surf | -28.12 | -46.17 | 355 | 18 | 0.25 | 0.37 | 23.0 | 36.9 | 95.17426 |
| 165 | 2 | 114 | 2013-11-19 | 21:40 | 5 | Surf | -28.65 | -44.99 | 728 | 226 | 0.29 | 0.28 | 22.4 | 36.4 | 76.31027 |
| Trichod.1 | 2 | Surf | -27.80 | -47.10 | 1002 | 194 | 83.77926 | ||||||||
| Trichod.2 | 2 | Surf | -27.80 | -47.10 | 744 | 206 | 78.31579 |
- ! Use the logical operators == != > >= < <= is.na()
Recap
Import and Export data
Select and create columns
Summarize data
Joining
Long vs. Wide format
Displaying tables
Next time: Data visualization (ggplot2)
- Understand the “grammar” of graphics
- Create exploratory graphics
Reading list
- Chapter 28 of R for data science
- Fundamental of data visualization
- Data visualization: practical introduction