Daniel Vaulot


Phytoplankton biogeography - metaPR2
Outline
Intro to metaPR2
Visualization/Analysis
MetaPR2 in practice
metaPR2: a database of metabarcodes
Metabarcoding
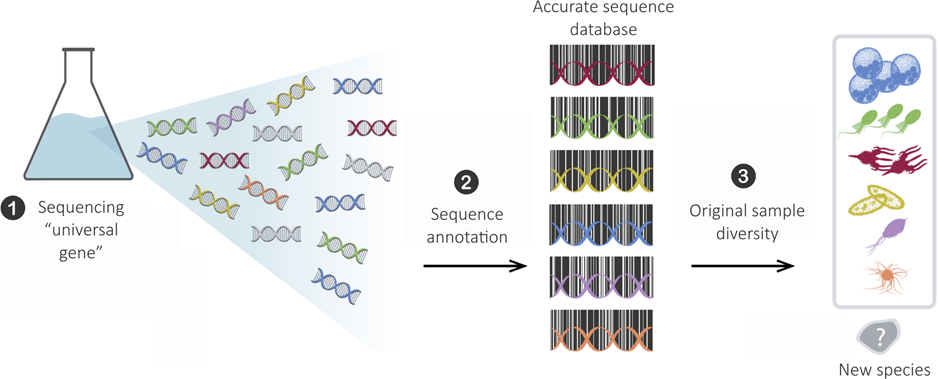
Many metabarcoding studies for eukaryotes

But hard to use…
- Processed with different pipelines
- Different primers
- Different levels of similarity
- Different reference databases
- Metadata lacking
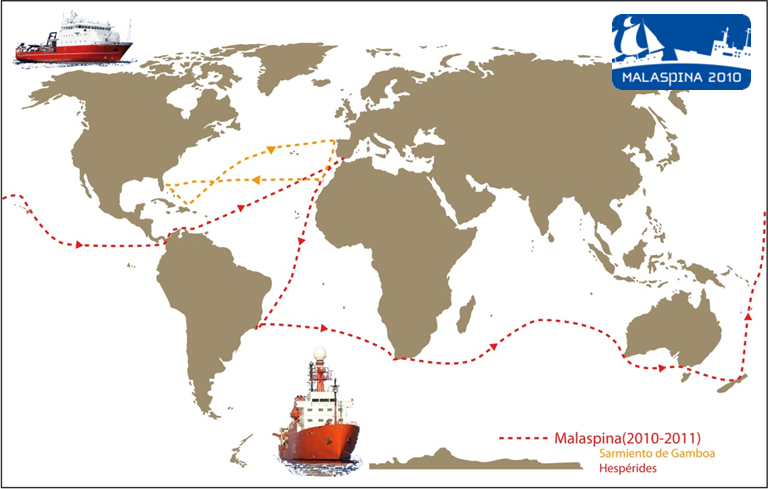
Large datasets
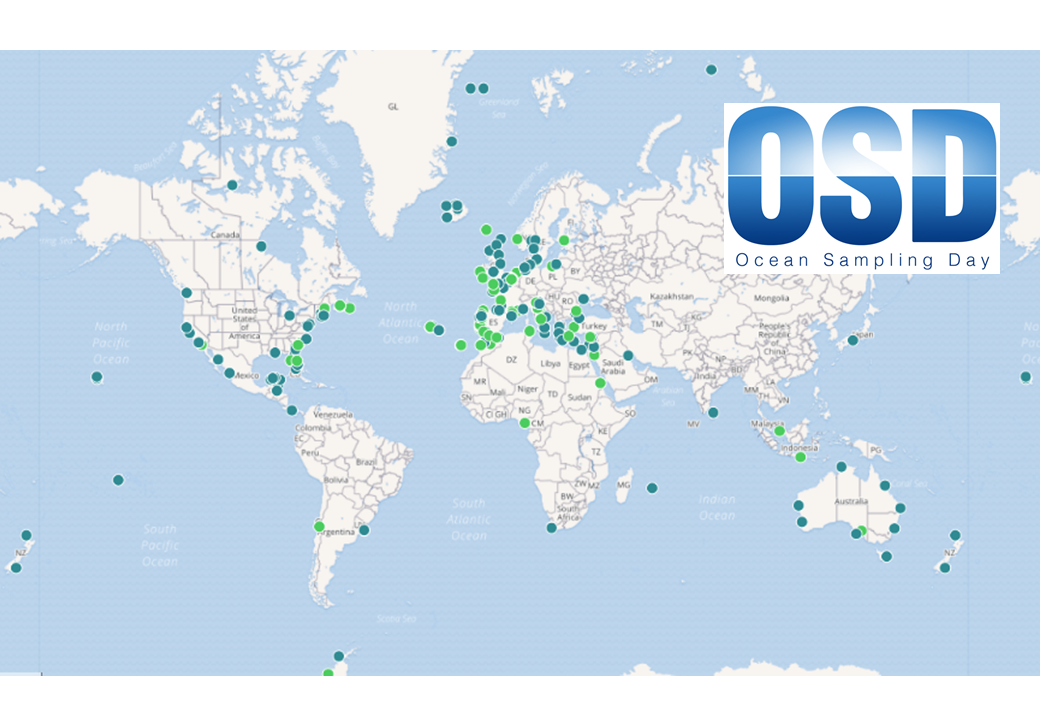
- Ocean Sampling Day
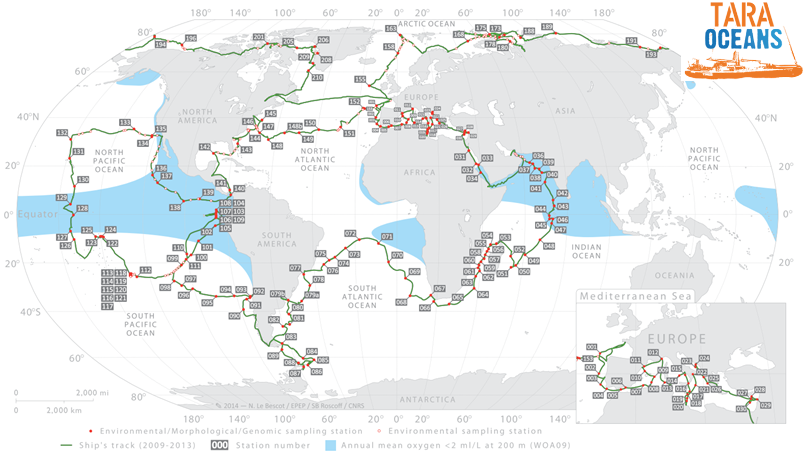
- Tara Oceans
- Malaspina
metaPR2 a database of metabarcodes
Reprocess public data
- Download Genbank (SRA) data
- Raw sequences
- Metadata
- Reprocess
- Amplicon Sequence Variant (dada2)
- Merge ASVs with same sequence
- Store in MySQL database
- Develop under R
- Web interface and R package
- https://app.metapr2.org
Current status
- Version 2.1
- Datasets: 59
- Samples: 6,202
- Barcodes (ASVs): 93,127
Factors affecting protist communities
Substrate
- Water
- Ice
- Sediment
- Soil
- Microbiome
Ecosystem
- Oceanic
- Coastal
- Rivers
- Lakes
- Terrestrial
Size fraction
- Total (0.2 µm -> 100 µm)
- Pico (0.2 µm -> 2-3 µm)
- Nano (2-3 µm -> 20 µm)
- Micro (20 µm -> 100-200 µm)
- Meso (100 µm -> 1000 µm)
Factors affecting protist communities
Environmental conditions
In oceanic waters:
- temperature
- salinity
- light
- nutrients
… which depend on:
- substrate (water vs.ice)
- latitude
- time of the year
- depth
- oceanic currents
- proximity of coast
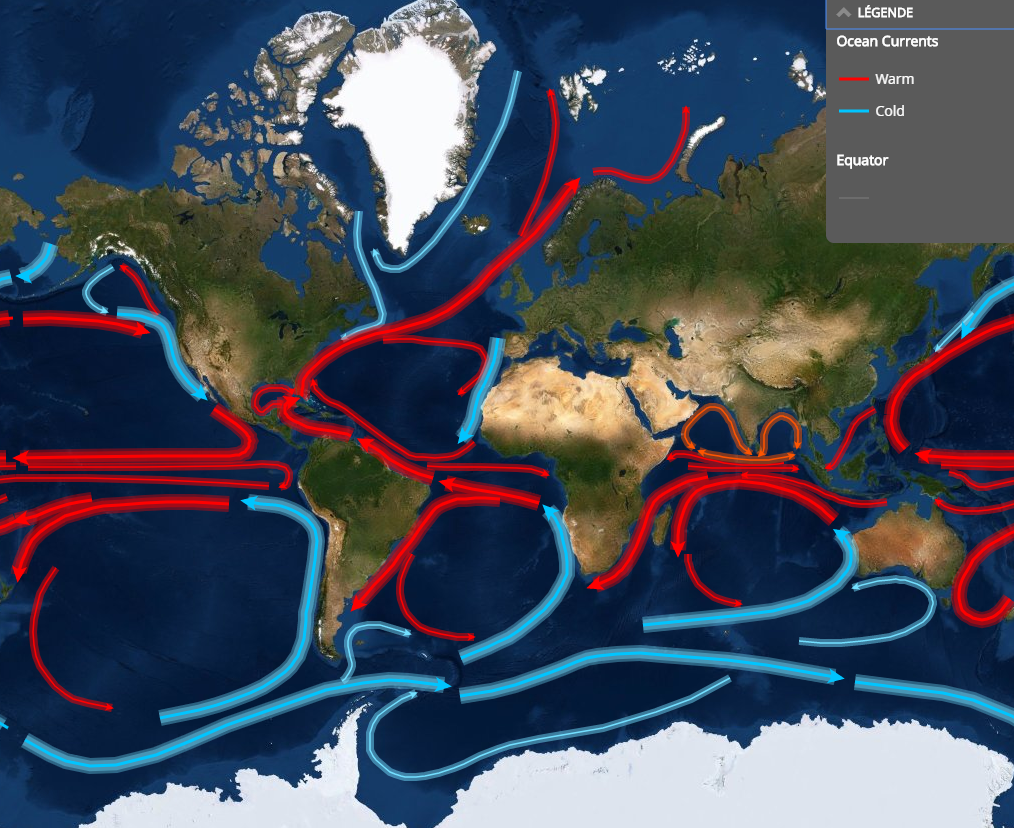
Example: Biogeography of Micromonas

Example: Biogeography of Ostreococcus

Metabarcoding pipeline
Overview
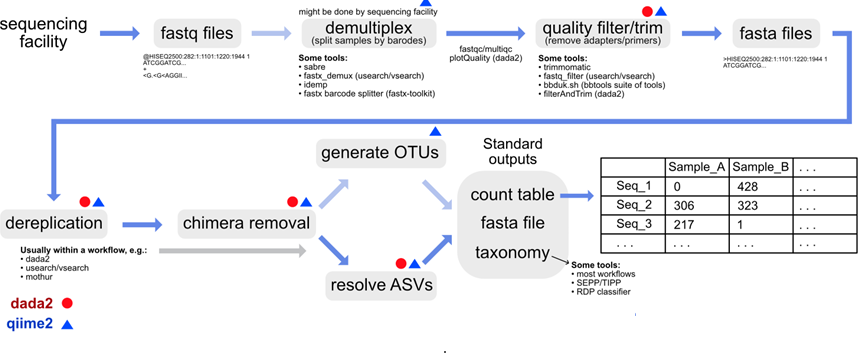
Sequences
Fastq files

Cluster
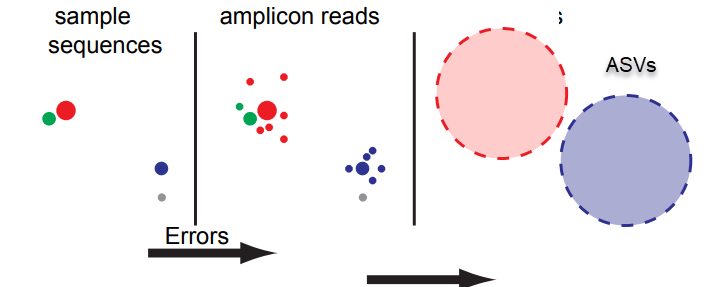
Assign
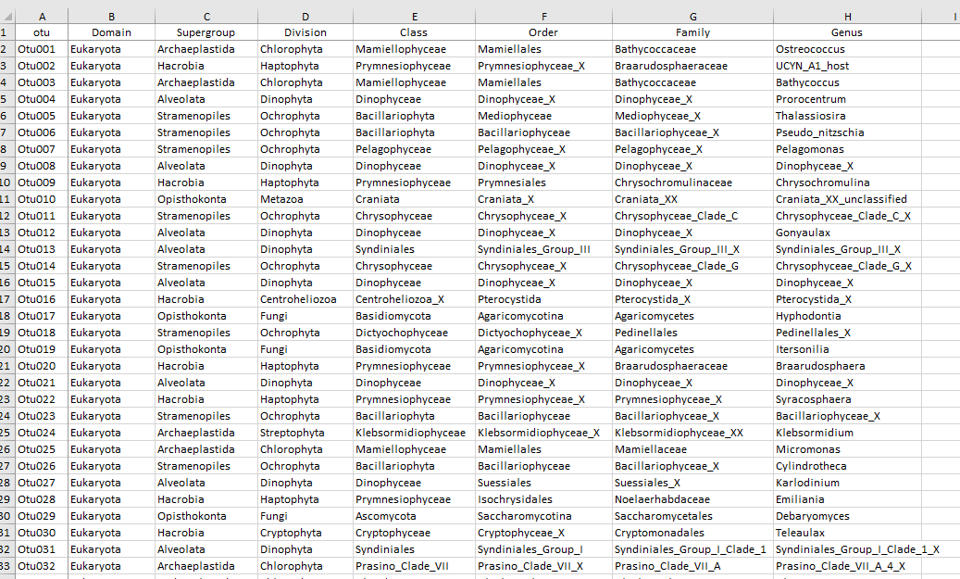
Output - ASVs
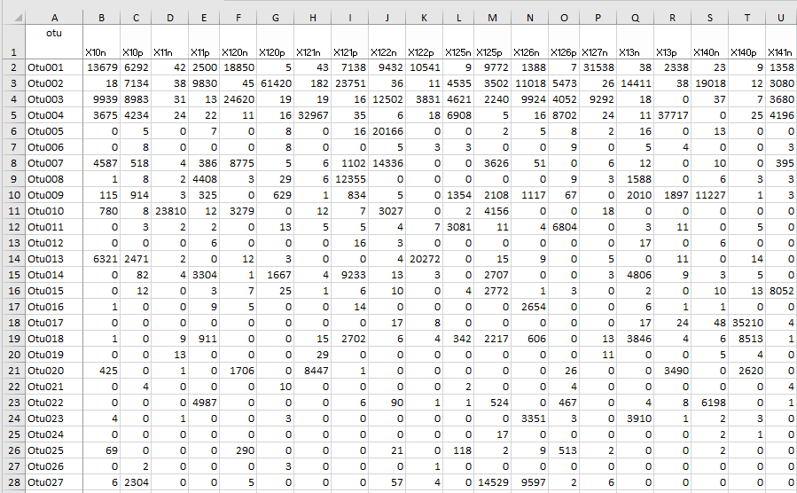
Output - Abundance
Output - Metadata
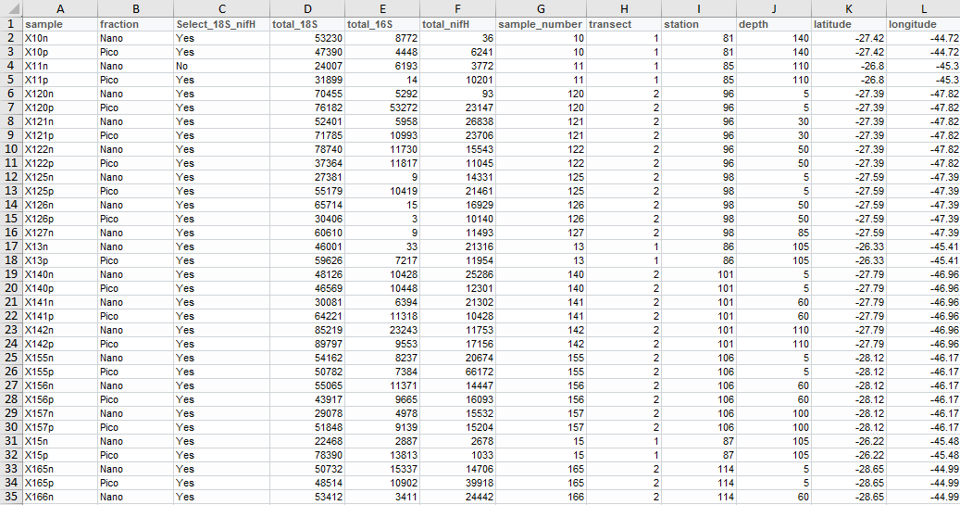
Output- Merged
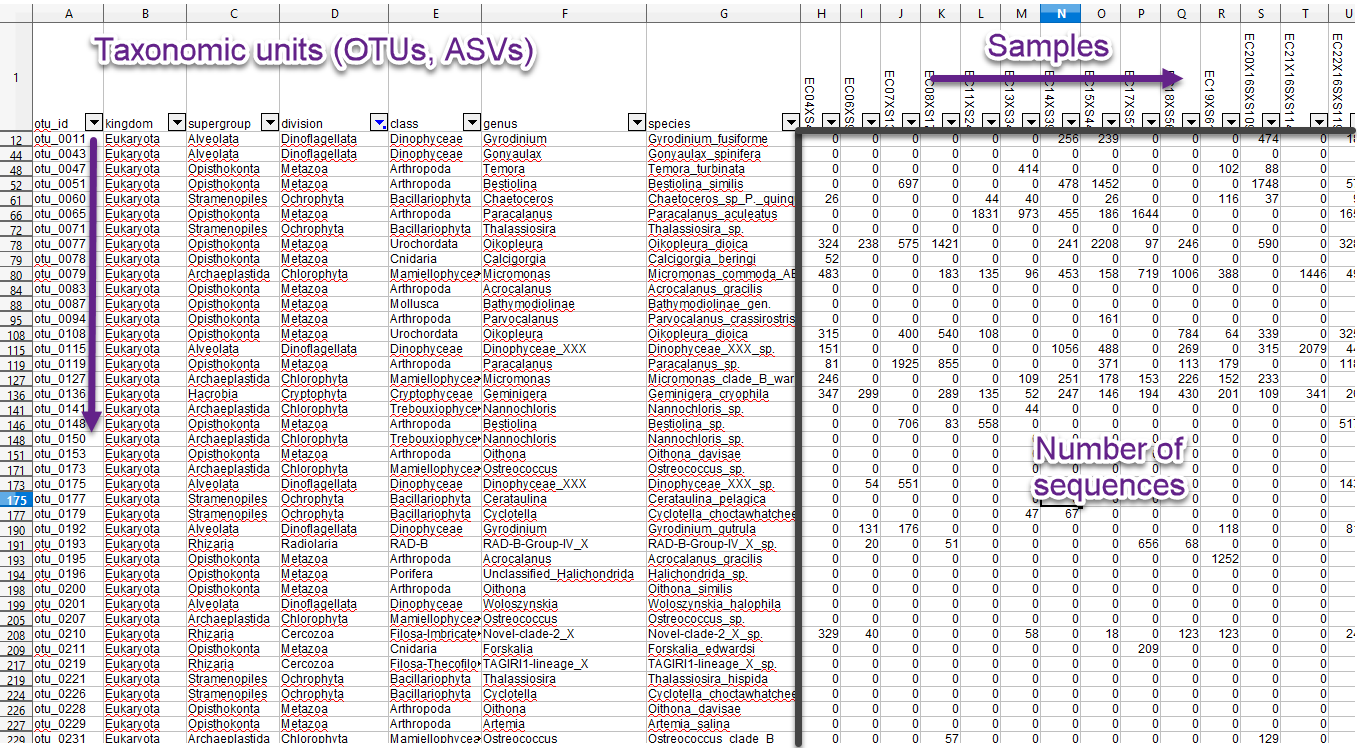
MetaPR2 - Main functions
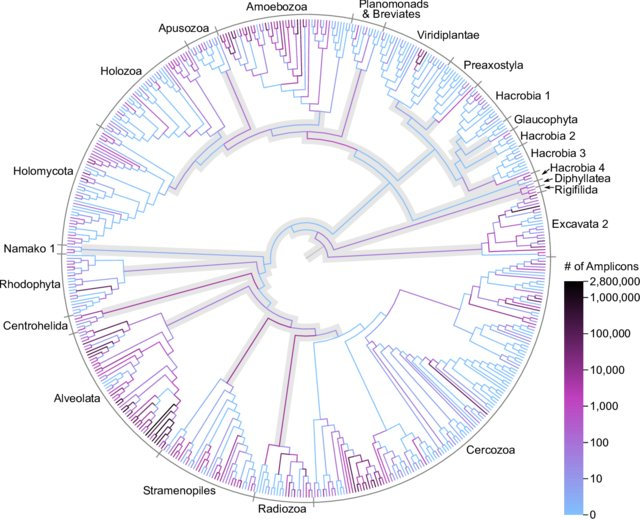
MetaPR2 - Taxonomy
Nine levels:
- Domain: Eukaryota
- Supergroup: Archaeplastida
- Division: Chlorophyta
- Subdivision: Chlorophyta_X
- Class: Mamiellophyceae
- Order: Mamielliales
- Family: Bathycoccaceae
- Genus: Bathycococcus
- Species: B. prasinos
Barplots - Latiude
Barplots - Time series

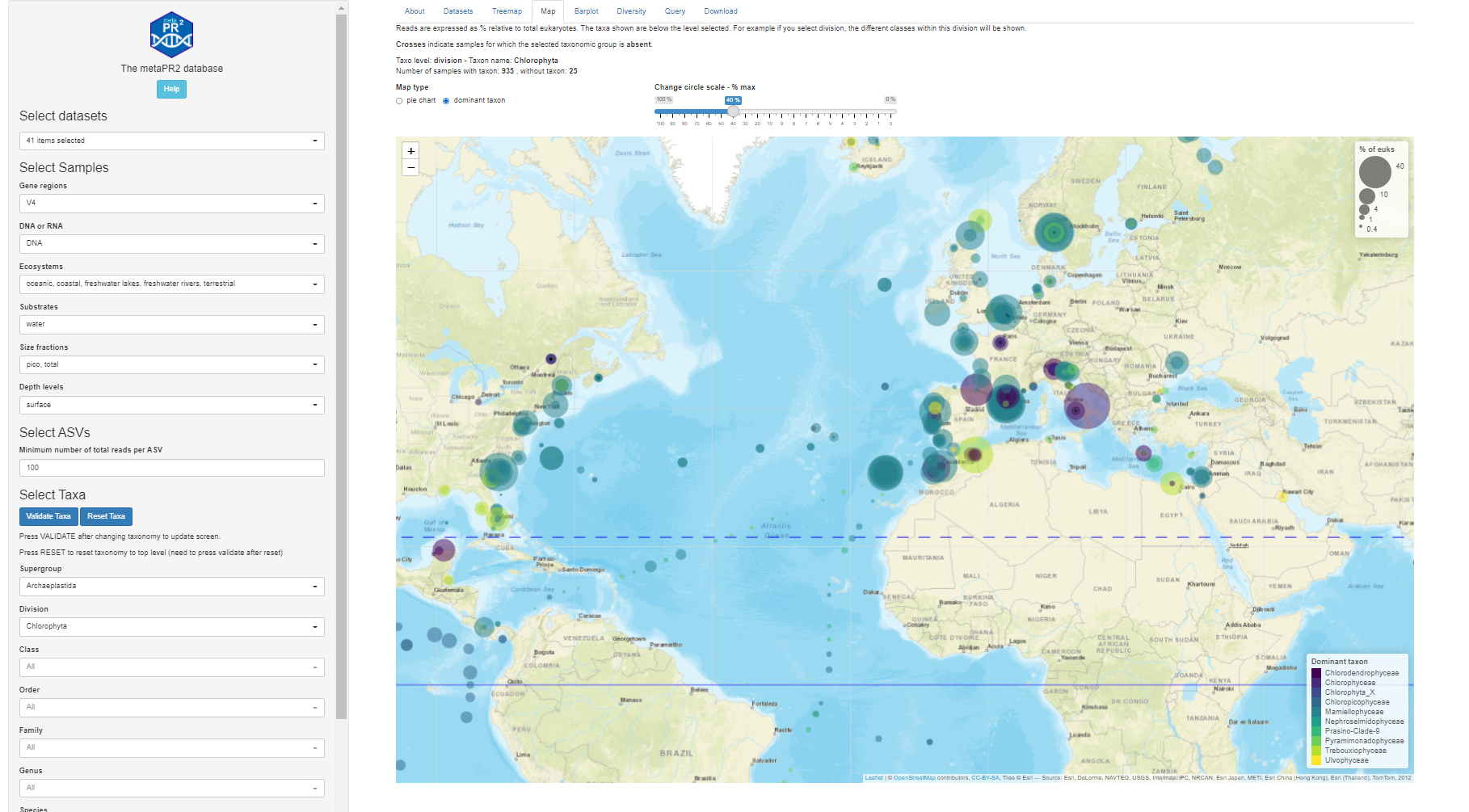
Maps - Dominant
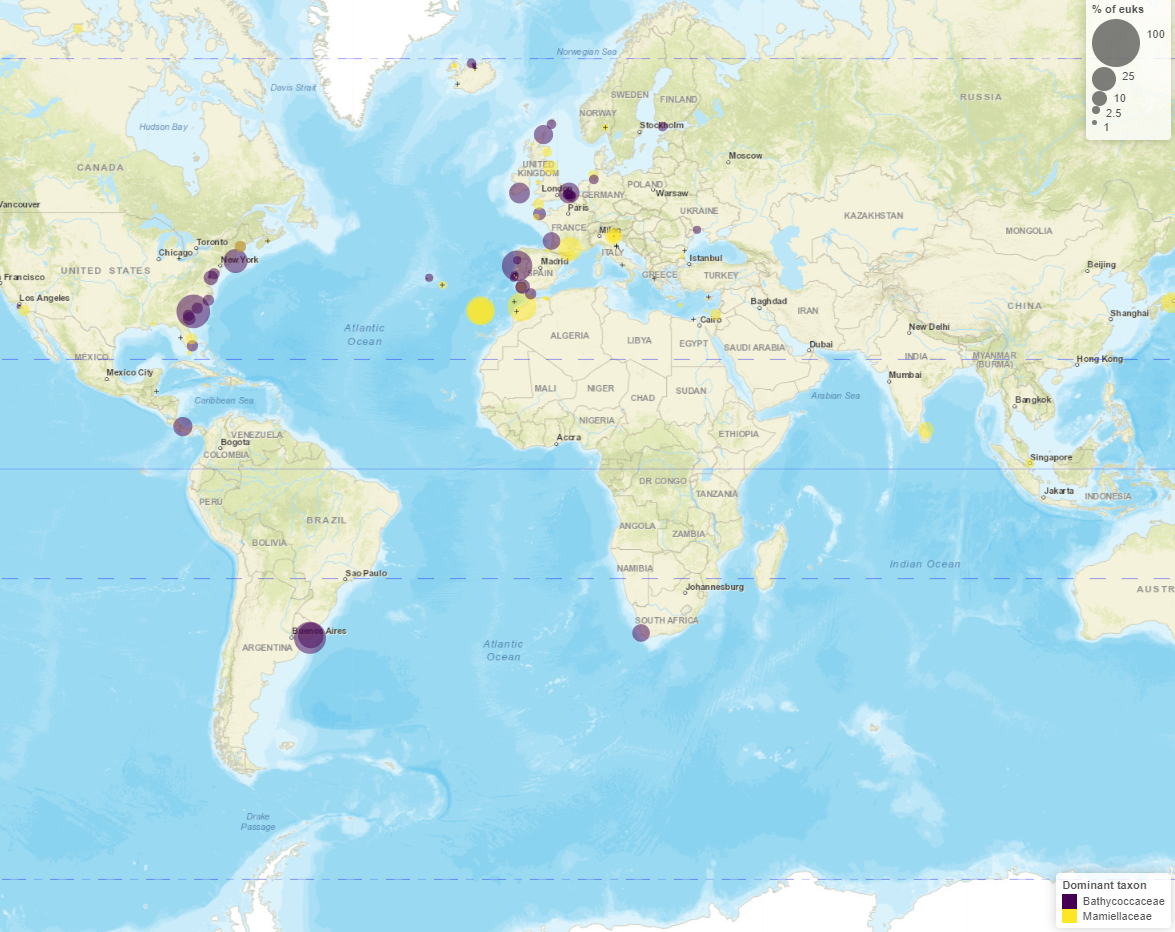
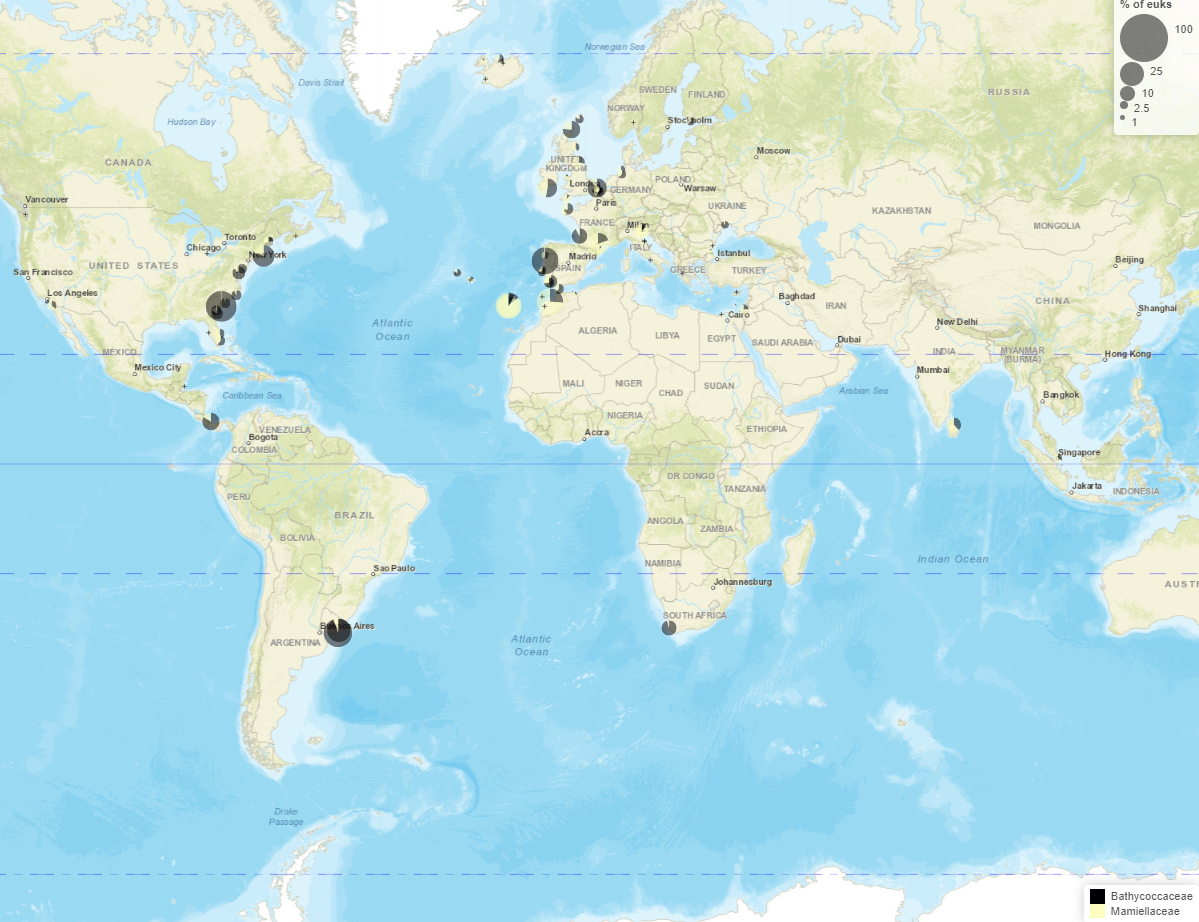
Maps - Pie charts
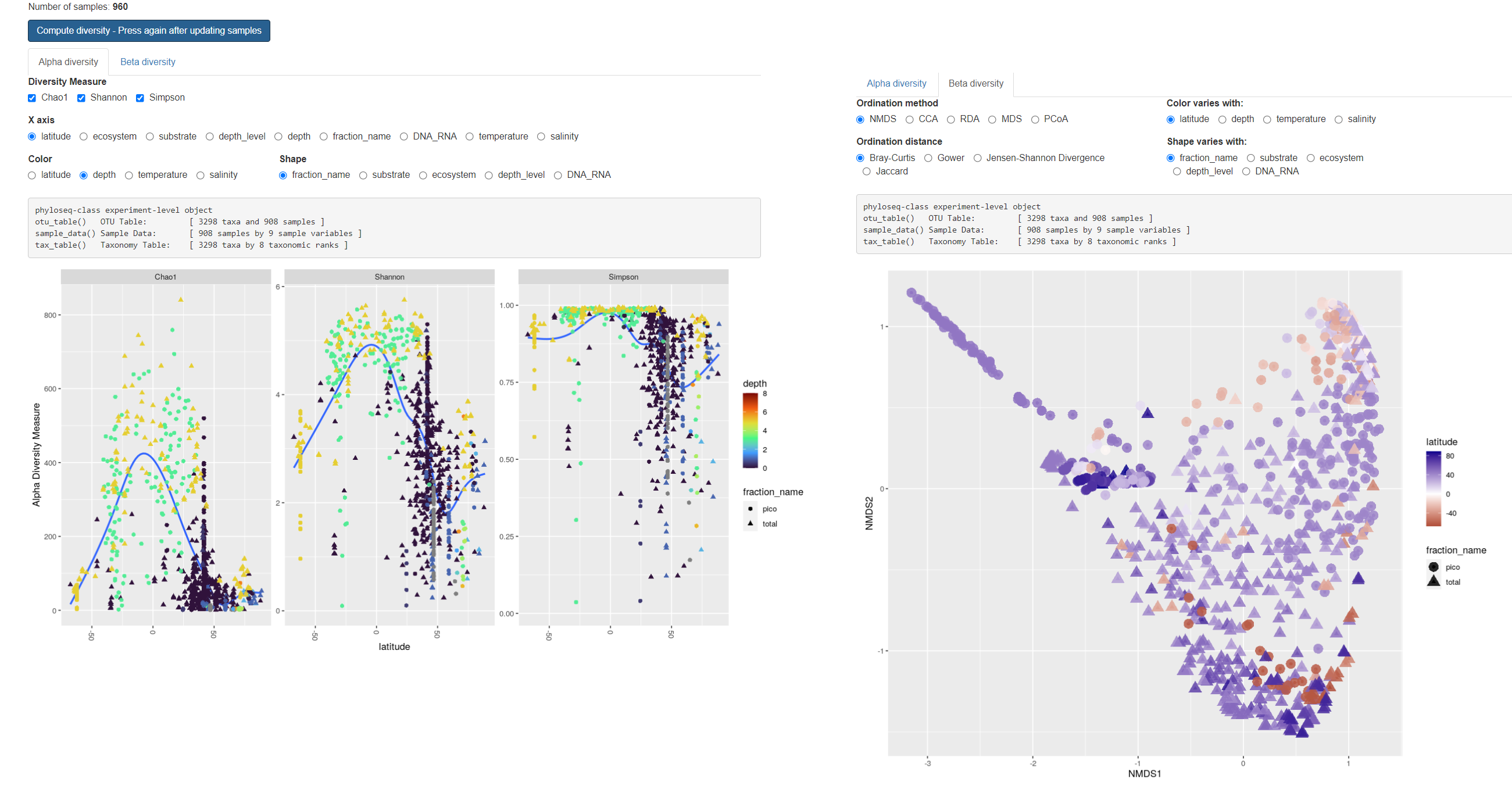
Diversity
MetaPR2 - In practice
Help and Samples
Help
- Read in detail
Sample table
- dataset_name
- paper (can be useful to read)
- number of samples
- number of ASVs
- number of reads per sample (coverage)
Sample selection
- Major datasets: OSD, Tara, Malaspina
- By habitat: oceanic, coastal etc…
- Start by “marine global V4”
- Extend to other habitats/datasets
- V4 vs V9
- DNA vs. RNA
- Ecosystems
- Sustrate: water, ice, soil…
- Size fractions: total, pico…
- Depth level: surface, euphotic…
- Minimum ASV: will filter out rare ASVs (e.g. 1000)
- Selection can be saved (yaml file)
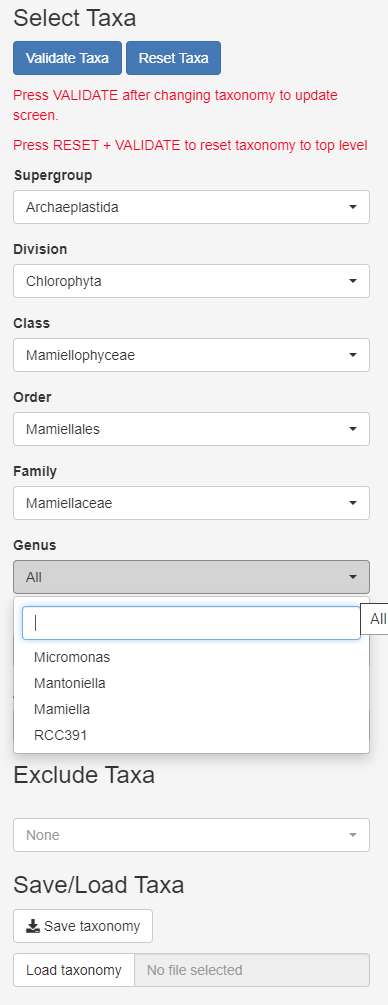
Taxonomy
- Can select several taxa within one level
- Press validate every time you need to refresh
- Can exclude taxa to remove fungi, metazoa…
- Can save taxonomy and reload taxonomy (yaml file)
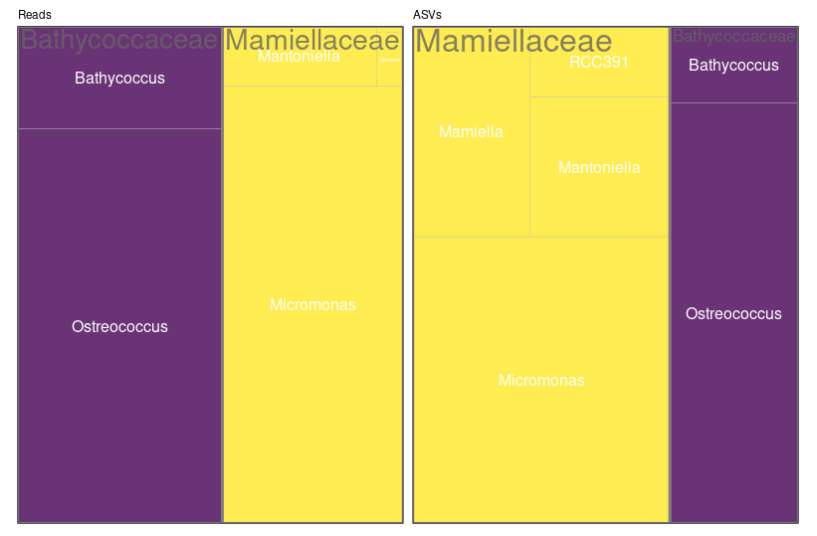
Treemaps, Maps and Barplots
Treemaps
- Left panel: abundance (number of reads)
- Reads are “normalized” to 100
- Right panel: diversity (number of ASVs)
Maps
- Read information at top
- Taxo level
- Number of samples with/without taxa
- Crosses where taxa absent
- Map types
- Dominant
- Pie chart
- Circle scale
- Moving right increases size
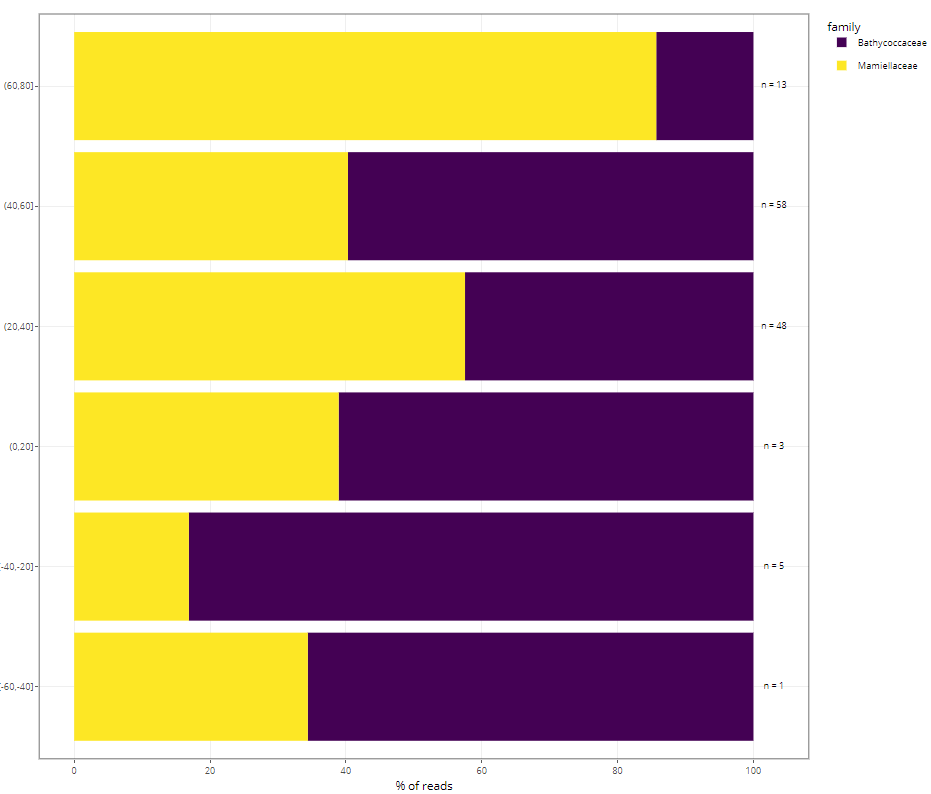
Barplots
- taxonomy vs. function
- variables to use (but this depends on samples selected !)
- fraction name
- ecosystem
- substrate
- depth level
- DNA_RNA
- latitude
- temperature
- salinity
- year, month, day for time series
Diversity
- Hit “Compute…” after refreshing taxonomy
- Time proportional to N samples and taxa
- Information about
- Number of samples
- Number of taxa (ASVs)
Alpha diversity
- X: Chao1, Shannon, Simpson (compare)
- Discretize continuous Y
- Change Y (see barplots)
- Change shape
- Change color
Beta diversity
- Ordination method (difference ?)
- Ordination distance (Bray, Jaccard…)
- Change color and shape
Download
- Download
- datasets (csv)
- samples (csv)
- asv list with taxonomy (csv)
- asv sequences (FASTA)
You can process these data with R (e.g. dplyr and ggplot2)
MetaPR2 home work
Green algae
- Prasinoderma
- Ostreococcus
Ochrophyta (Stramenopiles)
- Pelagomonas, Aureococcus
- Florenciella
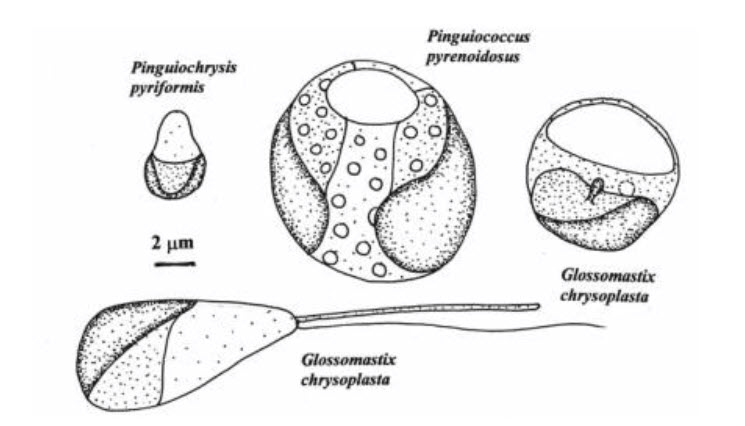
- Pinguiophyceae
Diatoms
- Pseudo-nitzschia
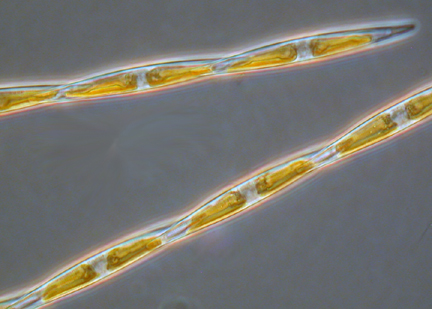
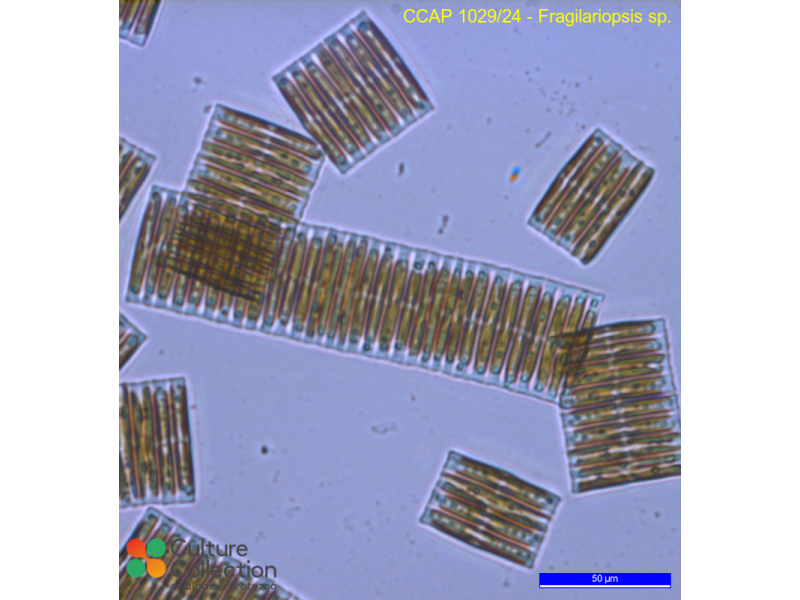
- Fragiliaropsis
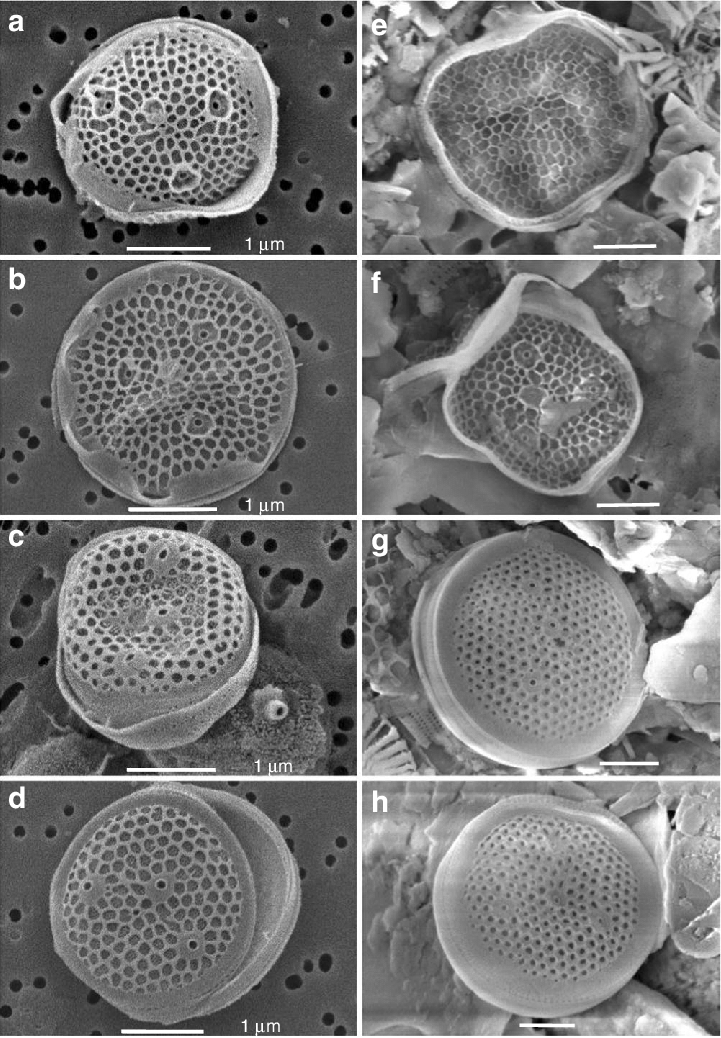
- Minidiscus
- Rhizosolenia
Dinoflagellates
- Dinophysis
- Ceratium, Tripos

Home work
Key points
- Look for key papers on this group
- What are the dominant species?
- What is the microdiversity [diversity within dominant species (ASVs)]?
- What is distribution ?
- Substrate (water, ice…)
- Ecosystems (marine, freshwater, terrestrial)
- Size fraction
- Depth layers (euphotic zone vs. meso and bathypelagic)
- Latitudinal bands (polar, temperate, tropical)
- Coastal vs Pelagic
- Alpha diversity
- Beta diversity
Final product (Optional)
- Use the proposal groups.
- Each group will have up to 10 pages to present their results.
- Structure as a paper.
- Use Quarto to write the paper (use template provided).
- Introduce very briefly the main biological characteristics and ecological importance of your taxonomic group.
- Explain which hypotheses/questions your group were interested in.
- Explain the results you have observed.